Diffusion Models in Medical Imaging: A Comprehensive Survey
-
摘要:
以扩散模型为代表的生成式人工智能近年来在医学成像领域取得了迅猛的进展。为了帮助更多学者全面的了解扩散模型这一先进技术,本文旨在提供扩散模型在医学成像领域的详细概述。首先以扩散模型的起源演变为主线介绍扩散建模框架的基础理论和基本概念;其次根据扩散模型的特点提供其在医学成像领域的系统分类,并涵盖不同成像模态如磁共振成像、计算机断层扫描、正电子发射计算机断层显像和光声成像等的广泛应用;最后讨论目前扩散模型的局限性并展望未来研究的潜在发展方向,为研究者后续的探索提供了一个直观的起始点。部分代码开源在网址:https://github.com/yqx7150/Diffusion-Models-for-Medical-Imaging。
Abstract:Generative artificial intelligence represented by diffusion models has significantly contributed to medical imaging reconstruction. To help researchers comprehensively understand the rich content of diffusion models, this review provides a detailed overview of diffusion models used in medical imaging reconstruction. The theoretical foundation and fundamental concepts underlying the diffusion modeling framework were first introduced, describing their origin and evolution. Second, a systematic characteristic-based taxonomy of diffusion models used in medical imaging reconstruction is provided, broadly covering their application to imaging modalities, including magnetic resonance imaging (MRI), computed tomography (CT), positron emission computed tomography (PET), and photoacoustic imaging (PAI). Finally, we discuss the limitations of current diffusion models and anticipate potential directions of future research, providing an intuitive starting point for subsequent exploratory research. Related codes are available at GitHub: https://github.com/yqx7150/Diffusion-Models-for-Medical-Imaging.
-
近年来,岩性复合型油气藏勘探已逐渐成为东海油气扩储增产的重要来源,但储集体普遍呈现低孔渗特征,流体引起的弹性差异微弱,空间非均质性展布规律显著。N构造位于西湖凹陷中央洼陷反转构造带中南部,紧邻区域生烃主洼,在轴向物源和西侧物源双源汇聚的背景下,受局部地貌和区域断裂条件影响,发育多种类型砂岩岩性体,周边已钻井揭示纵向发育多层系多套油气层。源-断-圈-储高效匹配具备有利油气成藏条件,开展致密砂岩储层流体描述,对于打开该构造背景上的岩性复合型油气藏勘探的突破口具有重要意义。
利用地震资料直接判断岩层含油气情况是国内外地球物理学家亟需攻克的热点课题[1]。相比于岩石固体骨架,孔隙流体所激发的地震信号微弱,通常借助地震波的运动学、动力学、形态学、弹性以及黏滞性等特征合理放大异常响应差异[2]。由此而产生了“亮点”技术、频率域吸收衰减属性以及流体因子等方法,其中以后者最为聚焦[3]。特别是随着振幅随偏移距的变化(amplitude variation with offset,AVO)技术的诞生和发展,基于叠前地震信息的流体描述技术得到了广泛关注和深入研究。
Ostrander[4]对岩心进行孔隙流体替换后发现叠前AVO响应特征存在明显差异;Chiburis[5]通过消除深浅层反射振幅能量差异,突出流体所引起的AVO响应,成功实现了叠前地震流体检测。Smith等[6]研究发现纵横波反射率的加权叠加能够有效区分流体,由此开启了利用“流体因子”进行油气检测的先河;Gidlow等[7]、Fatti等[8]和Wallace等[9]研究认为如何减弱背景因素对地震反射特征的影响是此类流体因子构建的关键,并给出了纵横波阻抗反射率的优化方案;Smith等[10]提出了“流体因子角”思想,与在阻抗域进行坐标旋转的泊松阻抗和扩展弹性阻抗理念相符;Goodway等[11-12]分析认为拉梅参数相比于岩石速度更具本质优势,实现了利用弹性模量开展烃类检测的跨越。此后,Hedlin[13]从岩石弹性模量中解耦出孔隙流体空间模量用以直接表征流体的弹性变化;Batzle[14]通过实验室测量发现利用体积模量与剪切模量的差值可进一步压制背景因素的影响提高流体识别能力;在双相多孔弹性介质理论的框架下,Russell等[15-16]对岩石流体项和固体项分别实现了数学形态解耦,发现流体项在时空域具有较高的流体描述精度。然而,研究对象的差异化导致每种方法均存在一定的局限性和适用性,必须结合实际工区优选高灵敏的烃检因子[17]。
叠前地震反演是从地震数据中提取岩石弹性信息的有效手段。苏世龙等[18]基于叠前纵横波阻抗同步反演开展储层含油气性预测;王保丽等[19]探讨了拉梅参数弹性阻抗方程的标准化形式,取得了较好的流体识别效果;宗兆云等[20]给出了流体项的纵横波模量表达形式,并利用弹性阻抗反演实现了流体敏感信息的直接提取;印兴耀等[21]深入解析了如何利用两项弹性阻抗反演提升深层 Russell流体因子可靠性的策略;杨培杰等[22]在贝叶斯理论框架下求解基于 Russell近似的AVO模型参数化方程实现了流体项的直接反演;邓炜等[23]以多孔弹性介质岩石物理模型为驱动,利用弹性阻抗反演实现了等效流体体积模量的直接反演,该方法对地震数据要求高,在致密储层存在求解不稳定问题;贾凌云等[24]结合广义弹性阻抗方程,利用新构建的敏感流体因子,对致密砂岩储层流体识别进行了尝试;周家雄等[25]将约束最小二乘与信赖域算法融合,通过对储层物性参数(粘土含量和孔隙度)的直接岩石物理反演进行“甜点”储层评价。但含水饱和度预测的收敛精度仍受制约;周林等[26]构建了精确佐普利兹方程的流体因子和泊松比的直接反演方法,提升了烃类检测精度和可靠性;Ma等[27]、Pan等[28]以岩石物理模型为驱动,探索了各向异性参数的直接反演方法。但预测结果对低频模型依赖度高,横向边界刻画不清晰。此外,传统叠前反演方法的初始模型导致地震振幅横向相对变化特征易被“模糊化”,并且地震子波对反演结果的影响程度较大[29]。
针对研究区致密砂岩储层地震流体识别所面临的岩石物理特征叠置严重、储层非均质性变化剧烈以及深层地震数据缺乏大角度信息等问题,本文结合岩石物理定性和定量判析技术,优选高灵敏烃检因子。以两项AVO模型参数化方程为基础,利用有色反演技术实现拉梅参数的直接提取,最终得到高灵敏烃检因子的地震流体敏感弹性信息。此成果应用于研究区对构造-岩性复合型气藏致密砂岩储层流体描述具有较高精度和适用性。
1. 方法原理
1.1 基于拉梅参数的两项AVO模型参数化方程
地震波反射系数的佐普利兹方程精确解及其近似式是对全弹性波方程的简化,为从叠前AVO信息中直接提取能够反映地层特性的弹性参数搭建了重要桥梁[30]。针对深层缺乏大角度信息的问题,刘晓晶等[31]推导了基于Russell近似的包含流体项f 和剪切模量μ的两项AVO近似式。当干岩石纵横波速度比的平方取为2.0时,就退化为包含拉梅模量λ和剪切模量μ的两项AVO近似式。可表示如下:
$$ \begin{aligned} {R_{{\rm{pp}}}}\left( \theta \right) = \,\Biggr( {\left( {\frac{1}{2} - \frac{1}{{\gamma _{{\rm{sat}}}^2}}} \right){{\sec }^2}\theta \frac{{\gamma _{{\rm{sat}}}^2{l_1} - 2{l_2}}}{{\left( {2{l_1} + 1} \right)\gamma _{{\rm{sat}}}^2 - 2\left( {2{l_2} + 1} \right)}}} \Biggr)\frac{{\Delta \lambda }}{\lambda } + \\ \Biggr( {\frac{1}{2} - \frac{2}{{\gamma _{{\rm{sat}}}^2}}{{\sin }^2}\theta - \left( {1 - \frac{1}{{\gamma _{{\rm{sat}}}^2}}{{\sec }^2}\theta } \right)\frac{{{l_2}}}{{1 + 2{l_2}}}} \Biggr)\frac{{\Delta \mu }}{\mu } \text{,}\; \end{aligned}$$ (1) 式中
$\gamma_{{\rm{sat}}} $ 表示饱和岩石纵横波速度比;l1和l2分别表示实际工区的密度与纵波速度、横波速度反射系数之间的拟合系数,θ表示阻抗差界面入射角度和透射角度之和的一半,λ和μ分别表示反射界面两侧拉梅参数模量和剪切模量的均值,Δλ和Δμ分别表示阻抗差界面相应拉梅参数模量和剪切模量的差值。表1为文献[26]中建立的双层砂岩模型弹性参数,其中关键参数
$\gamma ^2_{{\rm{dry}}} $ 和$\gamma ^2_{{\rm{sat}}} $ 的平均值分别取2.31和4.0。结合此模型开展佐普利兹方程精确解、Russell近似式和λ-μ近似式的反射系数精度对比分析。表 1 含气砂岩和含水砂岩模型弹性参数Table 1. Parameters of the sandstone model地层 Kdry/GPa μ/GPa $\gamma ^2_{{\rm{dry}}}$ $\gamma ^2_{{\rm{sat}}} $ $\rho $/(g/c3) VP/(m/s) VS/(m/s) 含水砂(上层) 3 3 2.32 4.48 1.9 2680 1265 含气砂(下层) 3 3 2.30 3.51 1.7 2520 1345 从图1中可以看到,λ-μ近似式与 Russell近似式的整体匹配度较高。特别地,在入射角小于20° 时两者与佐普利兹方程精确解的误差均较小;当入射角大于20° 时,相对误差均逐渐增大。因此,利用λ-μ近似式能够满足小角度入射条件下地震反演的精度需求。
1.2 叠前有色反演技术
有色反演技术是传统递归反演的改进算法,其本质思想是在频率域建立井间弹性参数与地震信息的映射关系,从而实现地震反射率体到弹性参数体的直接转换[32]。
地震记录利用褶积模型可数学表征为:
$$ s(t) = r(t) * w(t) \text{,} $$ (2) 式中:
$ s(t) $ 表示地震记录,$ r(t) $ 表示反射系数,$ w(t) $ 表示地震子波。将(2)式由时间域变换到频率域,即:
$$ S(\omega)=R(\omega) \cdot W(\omega) \text{,} $$ (3) 式中:
$ S(\omega ) $ 表示地震振幅谱,$ R(\omega ) $ 表示反射系数振幅谱,$ W(\omega ) $ 表示子波振幅谱。将(3)式两边取对数,得:
$$ \ln S(\omega)=\ln R(\omega)+\ln W(\omega) 。 $$ (4) 将界面反射系数变换到频率域,即:
$$ R(\omega)=\Big(Z_{2}(\omega)-Z_{1}(\omega)\Big) \Big/\Big(Z_{2}(\omega)+Z_{1}(\omega)\Big) 。 $$ (5) 将式(5)带入式(4)中,求得频率域匹配算子
$ W(\omega ) $ ,进一步通过傅里叶反变换即可获得时间域有色反演匹配算子$ w(t) $ 。该反演方法可有效减少稀井区井插值低频模型约束所造成的模型化问题,不仅能够保持反演结果在测井数据的量纲范围内波动,而且更好地反映了地震数据的细节特征。实现关键环节为[29]:①抽取靶区井旁地震道测井数据与地震数据分别进行振幅谱统计分析;②利用最小二乘拟合算法构建频率域匹配算子,进而转换得到时间域相位旋转 -90° 的有色算子;③结合上述算子将弹性反射率体直接转换为相对弹性数据体,开展无井反演;④通过频率域融合等算法,即可进一步实现层间绝对弹性数据体的有井反演。
1.3 拉梅参数弹性信息直接提取
叠前弹性参数直接反演技术是开展地震储层流体描述的重要手段,有效规避了间接计算的误差积累,具有更高的参数提取精度。利用(1)式建立叠前地震反射系数与拉梅模量λ和剪切模量μ在不同入射角情况下的映射关系:
$$ \left( {\begin{array}{*{20}{c}} \\ {{R_{PP}}({\theta _1},t)} \\ {{R_{PP}}({\theta _2},t)} \\ \vdots \\ {{R_{PP}}({\theta _n},t)} \\ \end{array}} \right) = \left( {\begin{array}{*{20}{c}} \\ {\begin{array}{*{20}{c}} {A({\theta _1},t)} \\ {A({\theta _2},t)} \\ \vdots \\ {A({\theta _n},t)} \\ \end{array}}&{\begin{array}{*{20}{c}} \\ {B({\theta _1},t)} \\ {B({\theta _2},t)} \\ \vdots \\ {B({\theta _n},t)} \end{array}}\\ \end{array}} \right)\left( {\begin{array}{*{20}{c}} \\ {\Delta \lambda /\lambda } \\ {\Delta \mu /\mu } \\ \end{array}} \right) \text{,} $$ (6) 式中
$ A\left( {{\theta _i},t} \right) $ 和$ B\left( {{\theta _i},t} \right) $ 分别表示方程系数,i表示地震数据不同采样点。通过反演算法求解上述矩阵方程组,即可从叠前道集中分别获得拉梅模量反射率
$ {{\Delta \lambda } \mathord{\left/ {\vphantom {{\Delta \lambda } \lambda }} \right. } \lambda } $ 和剪切模量反射率$ {{\Delta \mu } \mathord{\left/ {\vphantom {{\Delta \mu } \mu }} \right. } \mu } $ 的AVO属性体。进一步结合叠前有色反演直接获得相应的层间弹性信息,通过简单数学运算即可提取敏感流体因子弹性数据体[33]。关键实现操作流程,如图2所示。2. 实例应用
N构造位于西湖凹陷中央洼陷-反转构造带,是中新世晚期东西向挤压应力作用下形成的宽缓低幅背斜构造,属于典型的“洼中隆”。研究区发育缓坡背景下的河流-三角洲沉积体系,整体以三角洲平原分流河道砂体为主,主要目的层M组(M1、M2和M3)分布于3800~4100 m。受埋深压实作用、成岩作用和地质沉积环境影响,岩层岩石物理特征叠置严重,横向非均质性强,孔渗关系复杂。研究发现工区深层实际地震有效角度最大为25°;岩芯和壁芯资料表明储层孔隙度主要分布在0.8%~14.2%,平均值10.9%;渗透率主要分布在0.01~11.6 mD,平均值3.1 mD;总体为低孔低渗,局部呈特低孔特低渗特征。
实践证明该区存在低孔低渗背景下“一块多藏,一砂一藏”的构造-岩性复合型气藏模式,落实致密砂岩储层流体分布范围是打开该区新领域勘探突破的重要保障。本文利用拉梅参数直接反演技术实现了烃检因子地震流体弹性信息的直接提取,为致密砂岩储层含气性检测提供指导。
高质量的地震资料是开展深层致密储层烃类检测的有力保障。采用多步保真噪音压制技术有效压制涌浪噪音、线性及绕射噪音。针对浅水海域鬼波时空变剧烈问题,利用自适应参数优化方法实现震源及电缆鬼波的综合压制。通过τ-p域自相关扫描优化海底深度模型,实现水体速度校正和浅水多次波压制。数据规则化后,利用高精度 Radon变换压制残余多次波突出有效信号。基于波动方程建立包含振幅和相位的吸收补偿递推关系,实现叠前时间偏移道集的反Q补偿,提升地震资料分辨率。为后续地震流体识别奠定了坚实基础。
2.1 岩石物理敏感度判析
目前判别不同流体因子的识别能力主要采用岩石物理交会定性分析和数值化指标定量分析两种技术[34]。结合研究区已钻探井A井、B井和C井主力层的实测井数据以及解释成果,进行了地震岩石物理交会。从图3中可以看出,纵波阻抗ZP、横波阻抗ZS、体积密度ρ、剪切模量μ以及密度乘以剪切模量ρμ 对于含烃层和非烃层存在严重的叠置;泊松比σ、密度乘以拉梅模量ρλ、拉梅模量λ和扩展弹性阻抗 EEI 30° 对于含烃层和非烃层的区分度提升,但仍存在一定的叠置区间。而拉梅参数比值λ/μ的叠置区间最小,区分度最优。
流体敏感指示系数[35]通常作为衡量某种弹性参数对不同流体识别敏感度的定量判别标准,其数值越大对应识别灵敏度越高,如(7)式所示:
$$ H = \frac{{\left| {{E_{\rm{w}}} - {E_{\rm{g}}}} \right|}}{{{D_{\rm{g}}}}} \text{,} $$ (7) 其中,H表示流体敏感指示系数,其数值越大表示弹性参数对不同流体的区分能力越强;Dg表示储层含气时弹性参数的标准方差值;Ew表示储层含水时弹性参数的期望值;Eg表示储层含气时弹性参数的期望值。
这里对研究区12种弹性参数敏感度进行了统计计算。图4显示常规参数纵、横波阻抗和体积密度的敏感指示系数最低;泊松比、弹性模量及其密度组合和扩展弹性阻抗等整体敏感指示系数相对较高,其中剥离岩石骨架影响的拉梅参数比值灵敏度最高。因此,通过井间岩石物理判析表明拉梅参数比值
$ {\lambda \mathord{\left/ {\vphantom {\lambda \mu }} \right. } \mu } $ 能够有效满足对致密砂岩储层烃类检测的精度要求。2.2 弹性模量岩石物理量板分析
以实测数据为基础,结合Xu-White模型建立了拉梅参数的岩石物理量板。黏土含量变化范围为0%~50%,间隔25%;含水饱和度变化范围为0%~100%,间隔10%;孔隙度变化范围为0%~35%,间隔5%;其中流体相态混合模式采用更贴合实际的Brie经验公式。从图5中可以看到,剪切模量μ与孔隙度相关性较高,对流体的变化不敏感。在岩石高孔隙度时,拉梅模量λ能够有效识别流体;但随着孔隙度的减小,低孔气砂和高孔水砂弹性特征叠置严重,流体敏感性降低(图5(a))。而拉梅模量比值
$ {\lambda \mathord{\left/ {\vphantom {\lambda \mu }} \right. } \mu } $ 整体对于流体识别能力更强,叠置区间更小(图5(b))。因此,从岩石物理正演角度验证了该参数对致密储层流体识别的有效性。2.3 致密砂岩储层烃类检测
本研究区共有3口探井A井、B井和C井。主要目的层M组中M1层和M2层为低孔渗构造-岩性复合型气藏,M3层气饱含量低为含气水层。岩层阻抗差异小,叠后地震数据不能有效指示含烃层(图6(a))。本文分别开展常规扩展弹性阻抗反演(EEI 30°)和拉梅参数直接反演的烃类检测,并对比分析利用叠前同步反演间接估算拉梅参数比值
$ {\lambda \mathord{\left/ {\vphantom {\lambda \mu }} \right. } \mu } $ 的属性差异。图6(b)和图6(c)中红黄色代表低值含气性,蓝色代表高值含水性。在图中B井黑色虚方框处,M2层钻遇气层,M3层由两套相邻的含气水层砂体组成。在图中C井黑色虚椭圆框处,M1层揭示薄水层。对比可知,常规扩展弹性阻抗反演(EEI 30°)流体识别能力有限,目标区预测结果存在较强多解性。基于叠前同步反演间接计算的拉梅参数比值λ/μ 属性优于前者,但在黑色虚框处同样无法有效预测流体性质。主要是由于在缺乏大角度信息情况下利用三项式不能稳定提取弹性参数,并且间接计算会产生累计误差降低精度。而拉梅参数比值 λ/μ 直接反演结果清晰刻画了储集体内部流体的变化特征,整体与测井解释成果匹配度更高。主力层 M1层在该构造南部发育一条晚期南北向分流河道,切割北东向早期分流河道(图7)。地质油藏综合评价认为早期分流河道西南部发育构造-岩性复合型气藏,并且烃检存在强异常显示,设计部署了一口定向井。此外,工区南部为区域另一条河道主砂体发育部位,烃检异常明显(图7黑色实方框处),可作为后续该区滚动勘探开发的潜力目标区。
3. 总结与认识
为了落实研究区致密砂岩储层流体展布规律,本文引入一种基于拉梅参数直接反演的烃类检测方法。通过岩石物理定性和定量判析,确定拉梅参数比值对于研究区致密砂岩储层流体性质具有较高的识别能力。针对深层地震数据缺乏大角度信息的问题,采用两项AVO近似方程能够满足深层小角度入射的精度需求。充分利用叠前道集信息直接反演拉梅参数地震反射率数据体,并结合有色反演技术实现拉梅参数的直接估算,减小了间接估算的累计误差。有色反演技术有效保持了地震振幅的横向相对变化特征,使弹性参数地震提取更加客观。
该方法能够对研究区致密砂岩储层流体分布范围进行有效刻画,为后续勘探开发设计部署提供重要参考,于致密储层地震流体描述具有一定的应用价值和推广意义。
-
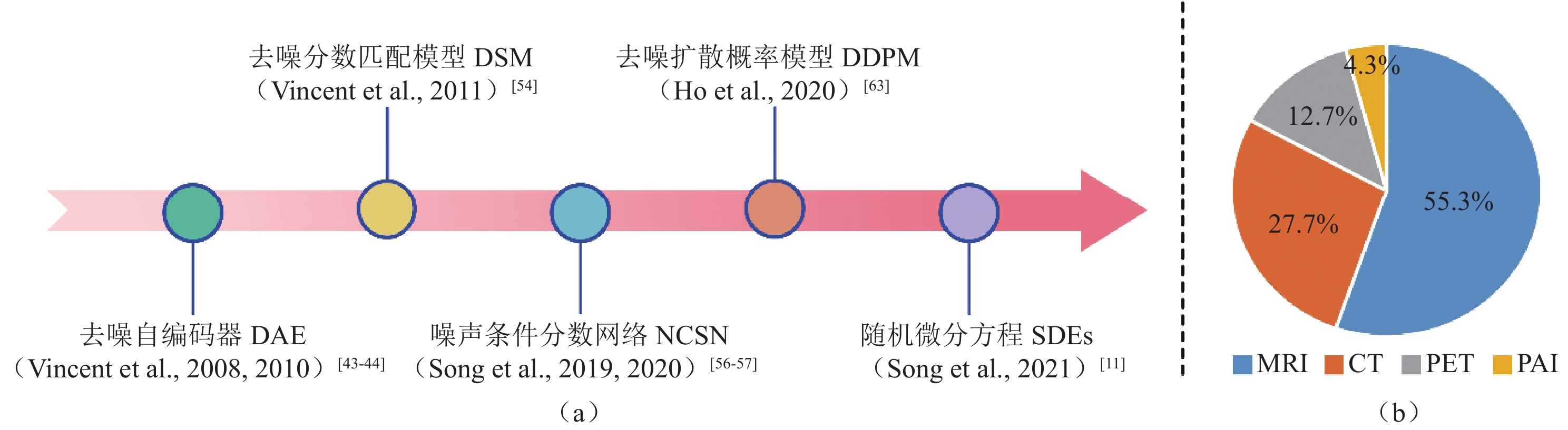
图 1 (a)扩散模型起源演变的时间轴;(b)按照关键词(diffusion model | medical imaging)、(score-based model | medical imaging)、(diffusion | medical | probabilistic model)在Google Scholar和Arxiv Sanity Preserver进行近5年相关论文检索,筛选并删除相同的结果最终统计扩散模型应用于不同成像模态分类的论文比例
Figure 1. (a) Timeline of the origin and evolution of the diffusion model; (b) Relevant papers published within the last five years found via Google Scholar and Arxiv Sanity Preserver searches using the keywords (diffusion model | medical imaging), (score-based model | medical imaging), and (diffusion | medical | probabilistic model). Duplicate results were identified and deleted before calculating the proportion of papers applying a diffusion model to different imaging modality classifications
图 3 扩散模型分别在图像域和K空间域的基本框架。其中前向过程从初始数据开始逐步添加噪声,使其最终接近纯噪声分布;逆向过程通过学习逐步去噪,从随机噪声中还原为高质量的目标数据
Figure 3. Basic framework of the diffusion model in the image and K-space domain. The forward process gradually adds noise from the initial data to approach a pure noise distribution; the reverse process gradually removes noise through learning and recovers high-quality target data from random noise
表 1 五大生成模型全方位对比
Table 1 Comprehensive comparison of five generative models
特点/模型 VAE GAN EBM Flow Diffusion Model 基本原理 由编码器将数据映射到潜在空间概率分布,再由解码器生成数据 生成器和判别器对抗训练,生成器生成数据,判别器区分真假 定义一个可微的能量函数,将数据点的概率分布与其能量值联系 通过可逆变换将简单分布映射到复杂数据分布。 从噪声分布起,逐步去噪恢复目标数据 优点 1.生成质量高
2.训练稳定
3.潜在空间连续1.生成质量高
2.多样性好
3.应用广泛1.灵活性高
2.隐式生成
3.表示能力强1.高效样本生成和密度估计
2.可解释性强1.生成质量高
2.强大的建模能力
3.广泛的应用场景缺点 1.生成样本模糊
2.计算复杂度高
3.难捕捉复杂分布1.训练困难
2.对数据敏感
3.计算资源消耗大1.配分函数难计算
2.训练不稳定
3.采样效率低1.设计合适的变换
2.模块具有挑战性
3.计算资源需求高1.训练过程复杂
2.对噪声模型依赖性
3.生成速度较慢训练稳定性 稳定 不稳定 稳定 稳定 稳定 生成质量 较高 高 较高 高 高 计算资源需求 中等 高 中等 高 高 模型复杂度 中等 高 中等 高 高 可解释性 较好 较差 较好 好 较差 灵活性 较低 高 高 高 高 对数据分布假设 较强 较弱 较强 较强 较弱 数据质量敏感性 较低 高 较低 较低 较低 生成速度 中等 中等 中等 中等 较慢 表 2 不同重建算法结果定量比较
Table 2 Quantitative comparison between the results of different reconstruction algorithms
ESPIRiT LINDBERG EBMRec SAKE WKGM SVD-WKGM T1 GE brain 2D random R=4 39.08/0.933 38.98/0.961 40.17/0.968 41.54/0.952 40.67/0.969 43.85/0.970 2D random R=6 36.01/0.921 35.16/0.958 36.55/0.952 38.09/0.932 37.14/0.957 39.94/0.960 T2 transverse brain 2D Poisson R=4 31.74/0.819 32.87/0.901 33.19/0.915 33.91/0.896 33.35/0.907 34.58/0.917 2D Poisson R=10 28.95/0.798 26.17/0.822 29.59/0.839 29.75/0.823 29.17/0.823 31.69/0.841 -
[1] BAO J M, CHEN D, WEN F, et al. CVAE-GAN: Fine-grained image generation through asymmetric training[C]//Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 2017: 2745-2754. DOI: 10.1109/iccv.2017.299.
[2] RAZAVI A, Van den OORD A, VINYALS O. Generating diverse high-fidelity images with VQ-VAE-2[J]. Advances in Neural Information Processing Systems, 2019, 32. DOI: 10.48550/arXiv.1906.00446.
[3] KONG Z F, PING W, HUANG J J, et al. Diffwave: A versatile diffusion model for audio synthesis[J]. arXiv Preprint arXiv: 2009.09761, 2020.
[4] OORD A, DIELEMAN S, ZEN H, et al. Wavenet: A generative model for raw audio[J]. arXiv Preprint arXiv: 1609.03499, 2016.
[5] LI X, THICKSTUN J, GULRAJANI I, et al. Diffusion-lm improves controllable text generation[J]. Advances in Neural Information Processing Systems, 2022, 35: 4328-4343. DOI: 10.48550/arXiv.2205.14217.
[6] YANG G D, HUANG X, HAO Z K, et al. Pointflow: 3D point cloud generation with continuous normalizing flows[C]//Proceedings of the IEEE/CVF international conference on computer vision, Seoul, Korea (South), 2019: 4541-4550. DOI: 10.48550/arXiv.1906.12320.
[7] BOND-TAYLOR S, LEACH A, LONG Y, et al. Deep generative modelling: A comparative review of VAEs, GANs, normalizing flows, energy-based and autoregressive models[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2021. DOI: 10.48550/arXiv.2103.04922.
[8] GOODFELLOW I, POUGET-ABADIE J, MIRZA M, et al. Generative adversarial nets[J]. Advances in Neural Information Processing Systems, 2014, 27. DOI: 10.48550/arXiv.1406.2661.
[9] JIMENEZ REZENDE D, MOHAMED S, WIERSTRA D. Stochastic backpropagation and approximate inference in deep generative models[J]. arXiv Preprint arXiv: 1401.4082, 2014.
[10] DINH L, SOHL-DICKSTEIN J, BENGIO S. Density estimation using real nvp[J]. arXiv Preprint arXiv: 1605.08803, 2016.
[11] SONG Y, SOHL-DICKSTEIN J, KINGMA D P, et al. Score-based generative modeling through stochastic differential equations[J]. arXiv Preprint arXiv: 2011.13456, 2020.
[12] KARRAS T, AITTALA M, AILA T, et al. Elucidating the design space of diffusion-based generative models[J]. Advances in Neural Information Processing Systems, 2022, 35: 26565-26577. DOI: 10.48550/arXiv.2206.00364.
[13] SOHL-DICKSTEIN J, WEISS E, MAHESWARANATHAN N, et al. Deep unsupervised learning using nonequilibrium thermodynamics[C]//International Conference on Machine Learning, Lille, France, 2015: 2256-2265. DOI: 10.48550/arXiv.1503.03585.
[14] SONG Y, DURKAN C, MURRAY I, et al. Maximum likelihood training of score-based diffusion models[J]. Advances in Neural Information Processing Systems, 2021, 34: 1415-1428. DOI: 10.48550/arXiv.2101.09258.
[15] SINHA A, SONG J M, MENG C L, et al. D2c: Diffusion-decoding models for few-shot conditional generation[J]. Advances in Neural Information Processing Systems, 2021, 34: 12533-12548. DOI: 10.48550/arXiv.2106.06819.
[16] VAHDAT A, KREIS K, KAUTZ J. Score-based generative modeling in latent space[J]. Advances in Neural Information Processing Systems, 2021, 34: 11287-11302. DOI: 10.48550/arXiv.2106.05931.
[17] SAHARIA C, HO J, CHAN W, et al. Image super-resolution via iterative refinement[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, 45(4): 4713-4726. DOI: 10.48550/arXiv.2104.07636.
[18] PANDEY K, MUKHERJEE A, RAI P, et al. Diffusevae: Efficient, controllable and high-fidelity generation from low-dimensional latents[J]. arXiv Preprint arXiv: 2201.00308, 2022.
[19] BAO F, LI C X, ZHU J, et al. Analytic-dpm: An analytic estimate of the optimal reverse variance in diffusion probabilistic models[J]. arXiv Preprint arXiv: 2201.06503, 2022.
[20] DOCKHORN T, VAHDAT A, KREIS K. Score-based generative modeling with critically-damped Langevin diffusion[J]. arXiv Preprint arXiv: 2112.07068, 2021.
[21] LIU N, LI S, DU Y L, et al. Compositional visual generation with composable diffusion models[C]//European Conference on Computer Vision, Tel-Aviv, Israel, 2022: 423-439. DOI: 10.48550/arXiv.2206.01714.
[22] JIANG Y M, YANG S, QIU H N, et al. Text2human: Text-driven controllable human image generation[J]. ACM Transactions on Graphics (TOG), 2022, 41(4): 1-11. DOI: 10.48550/arXiv.2205.15996.
[23] BATZOLIS G, STANCZUK J, SCHONLIEB C B, et al. Conditional image generation with score-based diffusion models[J]. arXiv Preprint arXiv: 2111.13606, 2021.
[24] DANIELS M, MAUNU T, HAND P. Score-based generative neural networks for large-scale optimal transport[J]. Advances in Neural Information Processing Systems, 2021, 34: 12955-12965. DOI: 10.48550/arXiv.2110.03237.
[25] CHUNG H, SIM B, YE J C. Come-closer-diffuse-faster: Accelerating conditional diffusion models for inverse problems through stochastic contraction[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 12413-12422. DOI: 10.48550/arXiv.2112.05146.
[26] KAWAR B, ELAD M, ERMON S, et al. Denoising diffusion restoration models[J]. Advances in Neural Information Processing Systems, 2022, 35: 23593-23606. DOI: 10.48550/arXiv.2201.11793.
[27] ESSER P, ROMBACH R, BLATTMANN A, et al. Imagebart: Bidirectional context with multinomial diffusion for autoregressive image synthesis[J]. Advances in Neural Information Processing Systems, 2021, 34: 3518-3532. DOI: 10.48550/arXiv.2108.08827.
[28] LUGMAYR A, DANELLJAN M, ROMERO A, et al. Repaint: Inpainting using denoising diffusion probabilistic models[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 11461-11471. DOI: 10.48550/arXiv.2201.09865.
[29] JING B, CORSO G, BERLINGHIERI R, et al. Subspace diffusion generative models[C]//European Conference on Computer Vision, Tel-Aviv, Israel, 2022: 274-289. DOI: 10.1007/978-3-031-20050-2_17.
[30] BARANCHUK D, RUBACHEV I, VOYNOV A, et al. Label-efficient semantic segmentation with diffusion models[J]. arXiv preprint arXiv: 2112.03126, 2021.
[31] GRAIKOS A, MALKIN N, JOJIC N, et al. Diffusion models as plug-and-play priors[J]. Advances in Neural Information Processing Systems, 2022, 35: 14715-14728. DOI: 10.48550/arXiv.2206.09012.
[32] WOLLEB J, SANDKÜHLER R, BIEDER F, et al. Diffusion models for implicit image segmentation ensembles[C]//International Conference on Medical Imaging with Deep Learning, New Orleans, USA, 2022: 1336-1348. DOI: 10.48550/arXiv.2112.03145.
[33] AMIT T, SHAHARBANY T, NACHMANI E, et al. Segdiff: Image segmentation with diffusion probabilistic models[J]. arXiv Preprint arXiv: 2112.00390, 2021.
[34] ZIMMERMANN R S, SCHOTT L, SONG Y, et al. Score-based generative classifiers[J]. arXiv Preprint arXiv: 2110.00473, 2021.
[35] PINAYA W H L, GRAHAM M S, GRAY R, et al. Fast unsupervised brain anomaly detection and segmentation with diffusion models[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention, New Orleans, USA, 2022: 705-714. DOI: 10.48550/arXiv.2206.03461.
[36] WOLLEB J, BIEDER F, SANDKUHLER R, et al. Diffusion models for medical anomaly detection[C]//International Conference on Medical image computing and computer-assisted intervention, New Orleans, USA, 2022: 35-45. DOI: 10.48550/arXiv.2203.04306.
[37] WYATT J, LEACH A, SCHMON S M, et al. Anoddpm: Anomaly detection with denoising diffusion probabilistic models using simplex noise[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 650-656. DOI: 10.1109/CVPRW56347.2022.00080.
[38] JALAL A, ARVINTE M, DARAS G, et al. Robust compressed sensing MRI with deep generative priors[J]. Advances in Neural Information Processing Systems, 2021, 34: 14938-14954. DOI: 10.48550/arXiv.2108.01368.
[39] CHUNG H, YE J C. Score-based diffusion models for accelerated MRI[J]. Medical Image Analysis, 2022, 80: 102479. DOI: 10.48550/arXiv.2110.05243.
[40] CAO H Q, TAN C, GAO Z Y, et al. A survey on generative diffusion model[J]. arXiv Preprint arXiv: 2209.02646, 2022.
[41] YANG L, ZHANG Z L, SONG Y, et al. Diffusion models: A comprehensive survey of methods and applications[J]. ACM Computing Surveys, 2023, 56(4): 1-39. DOI: 10.48550/arXiv.2209.00796.
[42] KAZEROUNI A, AGHDAM E K, HEIDARI M, et al. Diffusion models in medical imaging: A comprehensive survey[J]. Medical Image Analysis, 2023: 102846. DOI: 10.1016/jmedia.2023.102846.
[43] VINCENT P, LAROCHELLE H, BENGIO Y, et al. Extracting and composing robust features with denoising autoencoders[C]//Proceedings of the 25th international conference on Machine learning, Helsinki, Finland, 2008: 1096-1103. DOI: 10.1145/1390156.1390294.
[44] VINCENT P, LAROCHELLE H, LAJOIE I, et al. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion[J]. Journal of Machine Learning Research, 2010, 11(12). DOI: 10.5555/1756006.1953039.
[45] VINCENT P. A connection between score matching and denoising autoencoders[J]. Neural computation, 2011, 23(7): 1661-1674. DOI: 10.1162/NECO_a_00142.
[46] SONG Y, ERMON S. Generative modeling by estimating gradients of the data distribution[J]. Advances in Neural Information Processing Systems, 2019, 32. DOI: 10.48550/arXiv.1907.05600.
[47] SONG Y, GARG S, SHI J, et al. Sliced score matching: A scalable approach to density and score estimation[C]//Proceedings of The 35th Uncertainty in Artificial Intelligence Conference, Tel Aviv, Israel, 2020: 574-584. DOI: 10.48550/arXiv.1905.07088.
[48] HO J, JAIN A, ABBEEL P. Denoising diffusion probabilistic models[J]. Advances in Neural Information Processing Systems, 2020, 33: 6840-6851. DOI: 10.48550/arXiv.2006.11239.
[49] ERHAN D, COURVILLE A, BENGIO Y, et al. Why does unsupervised pre-training help deep learning?[C]//Proceedings of the 13th International Conference on Artificial Intelligence and Statistics, Pittsburgh, USA, 2010: 201-208.
[50] Le CUN Y, FOGELMAN-SOULIE F. Modeles connexionnistes de l’apprentissage[J]. Intellectica, 1987, 2(1): 114-143. DOI: 10.3406/intel.1987.1804.
[51] GALLINARI P, LECUN Y, THIRIA S, et al. Distributed associative memories: A comparison[M]. La Villette, Paris: Proceedings of Cognitiva 87, 1987.
[52] SEUNG H S. Learning continuous attractors in recurrent networks[J]. Advances in Neural Information Processing Systems, 1997, 10. DOI: 10.5555/3008904.3008997.
[53] TEZCAN K C, BAUMGARTNER C F, LUECHINGER R, et al. MR image reconstruction using deep density priors[J]. IEEE Transactions on Medical Imaging, 2018, 38(7): 1633-1642. DOI: 10.1109/TMI.2018.2887072.
[54] LIU Q G, YANG Q X, CHENG H T, et al. Highly under-sampled magnetic resonance imaging reconstruction using autoencoding priors[J]. Magnetic Resonance in Medicine, 2020, 83(1): 322-336. DOI: 10.1002/mrm.27921.
[55] WANG S Y, LV J J, HE Z N, et al. Denoising auto-encoding priors in undecimated wavelet domain for MR image reconstruction[J]. Neurocomputing, 2021, 437: 325-338. DOI: 10.1016/j.neucom.2020.09.086.
[56] LIU X S, ZHANG M H, LIU Q G, et al. Multi-contrast MR reconstruction with enhanced denoising autoencoder prior learning[C]//2020 IEEE 17th International Symposium on Biomedical Imaging, Iowa City, USA, 2020: 1-5. DOI: 10.1109/ISBI45749.2020.9098334.
[57] ZHANG M H, LI M T, ZHOU J J, et al. High-dimensional embedding network derived prior for compressive sensing MRI reconstruction[J]. Medical Image Analysis, 2020, 64: 101717. DOI: 10.1016/j.media.2020.101717.
[58] BLOCK A, MROUEH Y, RAKHLIN A. Generative modeling with denoising auto-encoders and Langevin sampling[J]. arXiv Preprint arXiv: 2002.00107, 2020.
[59] PARISI G. Correlation functions and computer simulations[J]. Nuclear Physics B, 1981, 180(3): 378-384. DOI: 10.1016/0550-3213(81)90056-0.
[60] GRENANDER U, MILLER M I. Representations of knowledge in complex systems[J]. Journal of the Royal Statistical Society: Series B (Methodological), 1994, 56(4): 549-581. DOI: 10.1111/j.2517-6161.1994.tb02000.x.
[61] QUAN C, ZHOU J J, ZHU Y Z, et al. Homotopic gradients of generative density priors for MR image reconstruction[J]. IEEE Transactions on Medical Imaging, 2021, 40(12): 3265-3278. DOI: 10.48550/arXiv.2008.06284.
[62] ZHU W Q, GUAN B, WANG S S, et al. Universal generative modeling for calibration-free parallel MR imaging[C]//2022 IEEE 19th International Symposium on Biomedical Imaging, Kolkata, India, 2022: 1-5. DOI: 10.1109/ISBI52829.2022.9761446.
[63] HE Z N, ZHANG Y K, GUAN Y, et al. Iterative reconstruction for low-dose CT using deep gradient priors of generative model[J]. IEEE Transactions on Radiation and Plasma Medical Sciences, 2022, 6(7): 741-754. DOI: 10.48550/arXiv.2009.12760.
[64] LYU Q, WANG G. Conversion between CT and MRI images using diffusion and score-matching models[J]. arXiv Preprint arXiv: 2209.12104, 2022.
[65] CHUNG H, LEE E S, YE J C. MR image denoising and super-resolution using regularized reverse diffusion[J]. IEEE Transactions on Medical Imaging, 2022, 42(4): 922-934. DOI: 10.48550/arXiv.2203.12621.
[66] MENG X X, GU Y N, PAN Y S, et al. A novel unified conditional score-based generative framework for multi-modal medical image completion[J]. arXiv Preprint arXiv: 2207.03430, 2022.
[67] LUO G, BLUMENTHAL M, HEIDE M, et al. Bayesian MRI reconstruction with joint uncertainty estimation using diffusion models[J]. Magnetic Resonance in Medicine, 2023, 90(1): 295-311. DOI: 10.1002/mrm.29624.
[68] PENG C, GUO P F, ZHOU S K, et al. Towards performant and reliable undersampled MR reconstruction via diffusion model sampling[C]// International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, Singapore, 2022: 623-633. DOI: 10.1007/978-3-031-16446-0_59.
[69] GUNGOR A, DAR S U H, OZTURK S, et al. Adaptive diffusion priors for accelerated MRI reconstruction[J]. Medical Image Analysis, 2023: 102872. DOI: 10.1016/j.media.2023.102872.
[70] XIE Y T, LI Q Z. Measurement-conditioned denoising diffusion probabilistic model for under-sampled medical image reconstruction[J]. arXiv Preprint arXiv: 2203.03623, 2022.
[71] OZTURKLER B, LIU C, ECKART B, et al. SMRD: Sure-based robust MRI reconstruction with diffusion models[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, Canada, 2023: 199-209. DOI: 10.1007/978-3-031-43898-1_20.
[72] MIRZA M U, DALMAZ O, BEDEL H A, et al. Learning fourier-constrained diffusion bridges for MRI reconstruction[J]. arXiv Preprint arXiv: 2308.01096, 2023.
[73] BIAN W Y, JANG A, LIU F. Diffusion modeling with domain-conditioned prior guidance for accelerated MRI and qMRI reconstruction[J]. arXiv Preprint arXiv: 2309.00783, 2023.
[74] KORKMAZ Y, CUKUR T, PATEL V M. Self-supervised MRI reconstruction with unrolled diffusion models[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, Canada, 2023: 491-501. DOI: 10.1007/978-3-031-43999-5_47.
[75] CHEN L X, TIAN X Y, WU J J, et al. JSMoCo: Joint coil sensitivity and motion correction in parallel MRI with a self-calibrating score-based diffusion model[J]. arXiv Preprint arXiv: 2310.09625, 2023.
[76] RAVULA S, LEVAC B, JALAL A, et al. Optimizing sampling patterns for compressed sensing MRI with diffusion generative models[J]. arXiv Preprint arXiv: 2306.03284, 2023.
[77] SONG Y, SHEN L Y, XING L, et al. Solving inverse problems in medical imaging with score-based generative models[J]. arXiv Preprint arXiv: 2111.08005, 2021.
[78] CHUNG H, SIM B, RYU D, et al. Improving diffusion models for inverse problems using manifold constraints[J]. arXiv Preprint arXiv: 2206.00941, 2022.
[79] TU Z J, JIANG C, GUAN Y, et al. K-space and image domain collaborative energy-based model for parallel MRI reconstruction[J]. Magnetic Resonance Imaging, 2023, 99: 110-122. DOI: 10.1016/j.mri.2023.02.004.
[80] PENG H, JIANG C, CHENG J, et al. One-shot generative prior in hankel-k-space for parallel imaging reconstruction[J]. IEEE Transactions on Medical Imaging, 2023. DOI: 10.1109/TMI.2023.3288219.
[81] YU C M, GUAN Y, KE Z W, et al. Universal generative modeling in dual domains for dynamic MRI[J]. NMR in Biomedicine, 2023, 36(12): e5011. DOI: 10.1002/nbm.5011.
[82] GUAN Y, YU C M, LU S Y, et al. Correlated and multi-frequency diffusion modeling for highly under-sampled MRI reconstruction[J]. arXiv Preprint arXiv: 2309.00853, 2023.
[83] ZHANG W, XIAO Z W, TAO H, et al. Low-rank tensor assisted K-space generative model for parallel imaging reconstruction[J]. Magnetic Resonance Imaging, 2023, 103: 198-207. DOI: 10.1016/j.mri.2023.07.004.
[84] CUI Z X, CAO CT, LIU S N, et al. Self-score: Self-supervised learning on score-based models for MRI reconstruction[J]. arXiv Preprint arXiv: 2209.00835, 2022.
[85] CUI Z X, LIU C C, FAN X H, et al. Physics-informed deep MRI: K-space interpolation meets heat diffusion[J]. IEEE Transactions on Medical Imaging, 2024, 43(10): 3503-3520. DOI: 10.1109/TMI.2024.3462988.
[86] CAO C T, CUI Z X, LIU S N, et al. High-frequency space diffusion models for accelerated MRI[J]. arXiv Preprint arXiv: 2208.05481, 2022.
[87] GUAN Y, YU C M, CUI Z X, et al. Correlated and multi-frequency diffusion modeling for highly under-sampled MRI reconstruction[J]. IEEE Transactions on Medical Imaging, 2024, 43(10): 3490-3502. DOI: 10.1109/TMI.2024.3381610.
[88] HUANG B, LU S Y, LIU Q G, et al. One-sample diffusion modeling in projection domain for low-dose CT imaging[J]. IEEE Transactions on Radiation and Plasma Medical Sciences, 2024. DOI: 10.1109/TRPMS.2024.3392248.
[89] WU W W, WANG Y Y. Data-iterative optimization score model for stable ultra-sparse-view CT reconstruction[J]. arXiv Preprint arXiv: 2308.14437, 2023.
[90] ZHANG W H, HUANG B, LIU Q G, et al. Low-rank angular prior guided multi-diffusion model for few-shot low-dose CT reconstruction[J]. IEEE Transactions on Radiation and Plasma Medical Sciences, 2024. DOI: 10.1109/TCI.2024.3503366.
[91] LIU J M, ANIRUDH R, THIAGARAJAN J J, et al. DOLCE: A model-based probabilistic diffusion framework for limited-angle CT reconstruction[C]//Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2023: 10498-10508. DOI: 10.48550/arXiv.2211.12340.
[92] XIA W J, SHI Y Y, NIU C, et al. Diffusion prior regularized iterative reconstruction for low-dose CT[J]. arXiv Preprint arXiv: 2310.06949, 2023.
[93] XIA W J, CONG W X, WANG G. Patch-based denoising diffusion probabilistic model for sparse-view CT reconstruction[J]. arXiv Preprint arXiv: 2211.10388, 2022.
[94] XIA W J, NIU C, CONG W X, et al. Sub-volume-based denoising diffusion probabilistic model for cone-beam CT reconstruction from incomplete data[J]. arXiv e-Prints, 2023: arXiv: 2303.12861.
[95] GAO Q, LI Z L, ZHANG J P, et al. CoreDiff: Contextual error-modulated generalized diffusion model for low-dose CT denoising and generalization[J]. arXiv Preprint arXiv: 2304.01814, 2023.
[96] WU W W, WANG Y Y, LIU Q G, et al. Wavelet-improved score-based generative model for medical imaging[J]. IEEE Transactions on Medical Imaging, 2023. DOI: 10.1109/TMI.2023.3325824.
[97] XU K, LU S Y, HUANG B, et al. Stage-by-stage wavelet optimization refinement diffusion model for sparse-view CT reconstruction[J]. arXiv Preprint arXiv: 2308.15942, 2023.
[98] ZHANG J J, MAO H Y, WANG X R, et al. Wavelet-inspired multi-channel score-based model for limited-angle CT reconstruction[J]. IEEE Transactions on Medical Imaging, 2024, 43(10): 3436-3448. DOI: 10.1109/TMI.2024.3367167.
[99] GUAN B, YANG C L, ZHANG L, et al. Generative modeling in sinogram domain for sparse-view CT reconstruction[J]. IEEE Transactions on Radiation and Plasma Medical Sciences, 2023. DOI: 10.1109/TRPMS.2023.3309474.
[100] LIU X, XIE Y Q, DIAO S H, et al. Unsupervised CT metal artifact reduction by plugging diffusion priors in dual domains[J]. IEEE Transactions on Medical Imaging, 2024, 43(10): 3533-3545. DOI: 10.1109/TMI.2024.3351201.
[101] WANG Y Y, LI Z R, WU W W. Time-reversion fast-sampling score-based model for limited-angle CT reconstruction[J]. IEEE Transactions on Medical Imaging, 2024, 43(10): 3449-3460. DOI: 10.1109/TMI.2024.3418838.
[102] WU W W, PAN J Y, WANG Y Y, et al. Multi-channel optimization generative model for stable ultra-sparse-view CT reconstruction[J]. IEEE Transactions on Medical Imaging, 2024, 43(10): 3416-3475. DOI: 10.1109/TMI.2024.3376414.
[103] HAN Z Y, WANG Y H, ZHOU L P, et al. Contrastive diffusion model with auxiliary guidance for coarse-to-fine PET reconstruction[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, Canada, 2023: 239-249. DOI: 10.1007/978-3-031-43999-5_23.
[104] JIANG C W, PAN Y S, LIU M X, et al. PET-diffusion: Unsupervised PET enhancement based on the latent diffusion model[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, Canada, 2023: 3-12. DOI: 10.1007/978-3-031-43907-0_1.
[105] XIE T F, CAO C T, CUI Z X, et al. Brain pet synthesis from MRI using joint probability distribution of diffusion model at ultrahigh fields[J]. arXiv Preprint arXiv: 2211.08901, 2022.
[106] XIE T F, CAO C R, CUI Z X, et al. Synthesizing PET images from high-field and ultra-high-field MR images using joint diffusion attention model[J]. arXiv Preprint arXiv: 2305.03901, 2023.
[107] XIE T F, CUI Z X, LUO C, et al. Joint diffusion: Mutual consistency-driven diffusion model for PET-MRI co-reconstruction[J]. arXiv Preprint arXiv: 2311.14473, 2023.
[108] PAN S Y, ABOUEI E, PENG J B, et al. Full-dose PET synthesis from low-dose PET using high-efficiency diffusion denoising probabilistic model[J]. arXiv Preprint arXiv: 2308.13072, 2023.
[109] SONG X L, WANG G J, ZHONG W H, et al. Sparse-view reconstruction for photoacoustic tomography com-bining diffusion model with model-based iteration[J]. Photoacoustics, 2023, 33: 100558. DOI: 10.1016/j.pacs.2023.100558.
[110] TONG S Q, LAN H R, NIE L M, et al. Score-based generative models for photoacoustic image reconstruction with rotation consistency constraints[J]. arXiv Preprint arXiv: 2306.13843, 2023.
[111] DOSOVITSKIY A, BEYER L, KOLESNIKOV A, et al. An image is worth 16x16 words: Transformers for image recognition at scale[J]. arXiv Preprint arXiv: 2010.11929, 2020.
[112] WANG Z H, YANG Y Y, CHEN Y Z, et al. Mutual information guided diffusion for zero-shot cross-modality medical image translation[J]. IEEE Transactions on Medical Imaging, 2024, 43(8): 2825-2838. DOI: 10.1109/TMI.2024.3382043.
[113] CHU J Y, DU C H, LIN X Y, et al. Highly accelerated MRI via implicit neural representation guided posterior sampling of diffusion models[J]. Medical Image Analysis, 2025, 100.







 下载:
下载: