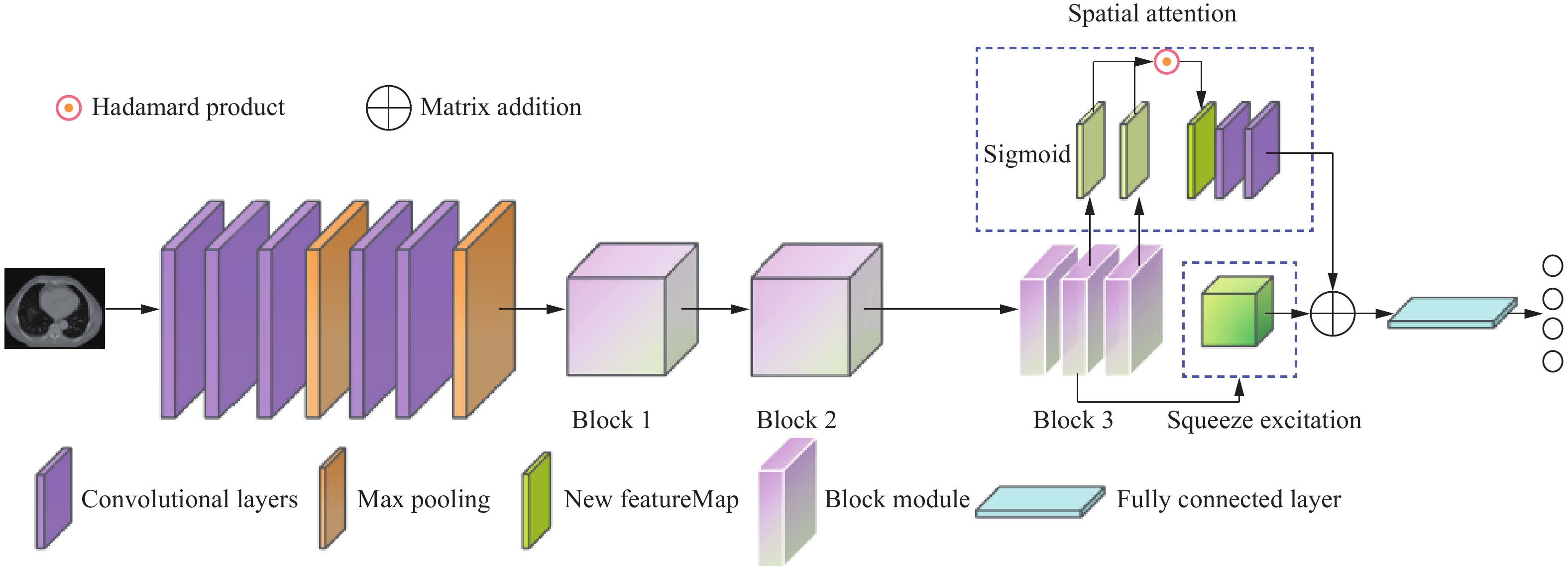
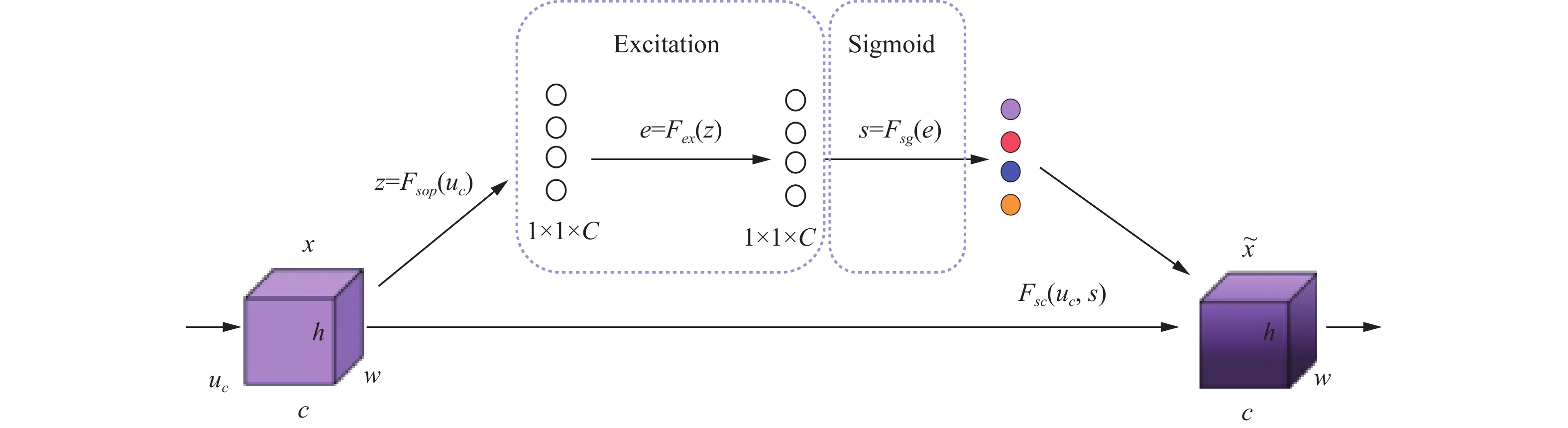
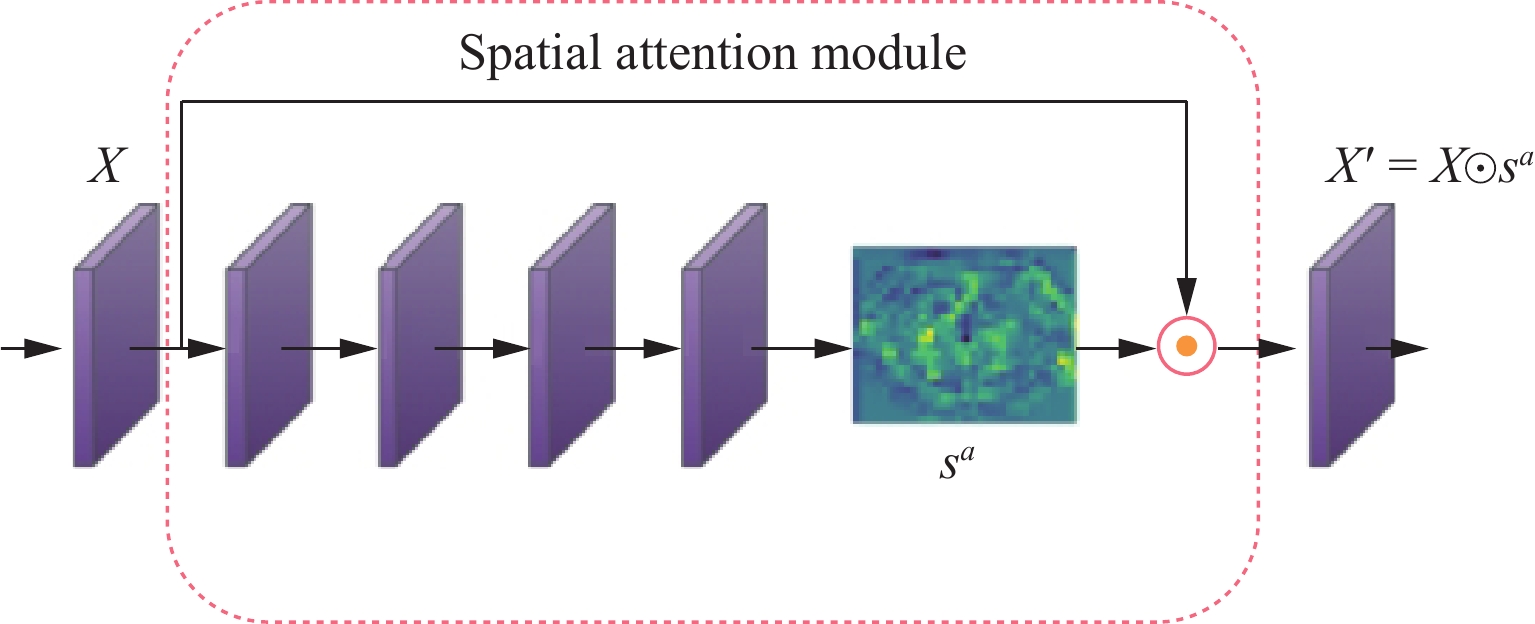
Deep Learning Computer-aided Diagnostic Model for Non-small Cell Lung Cancer Based on Convolutional Neural Network and Attention Mechanism
-
摘要:
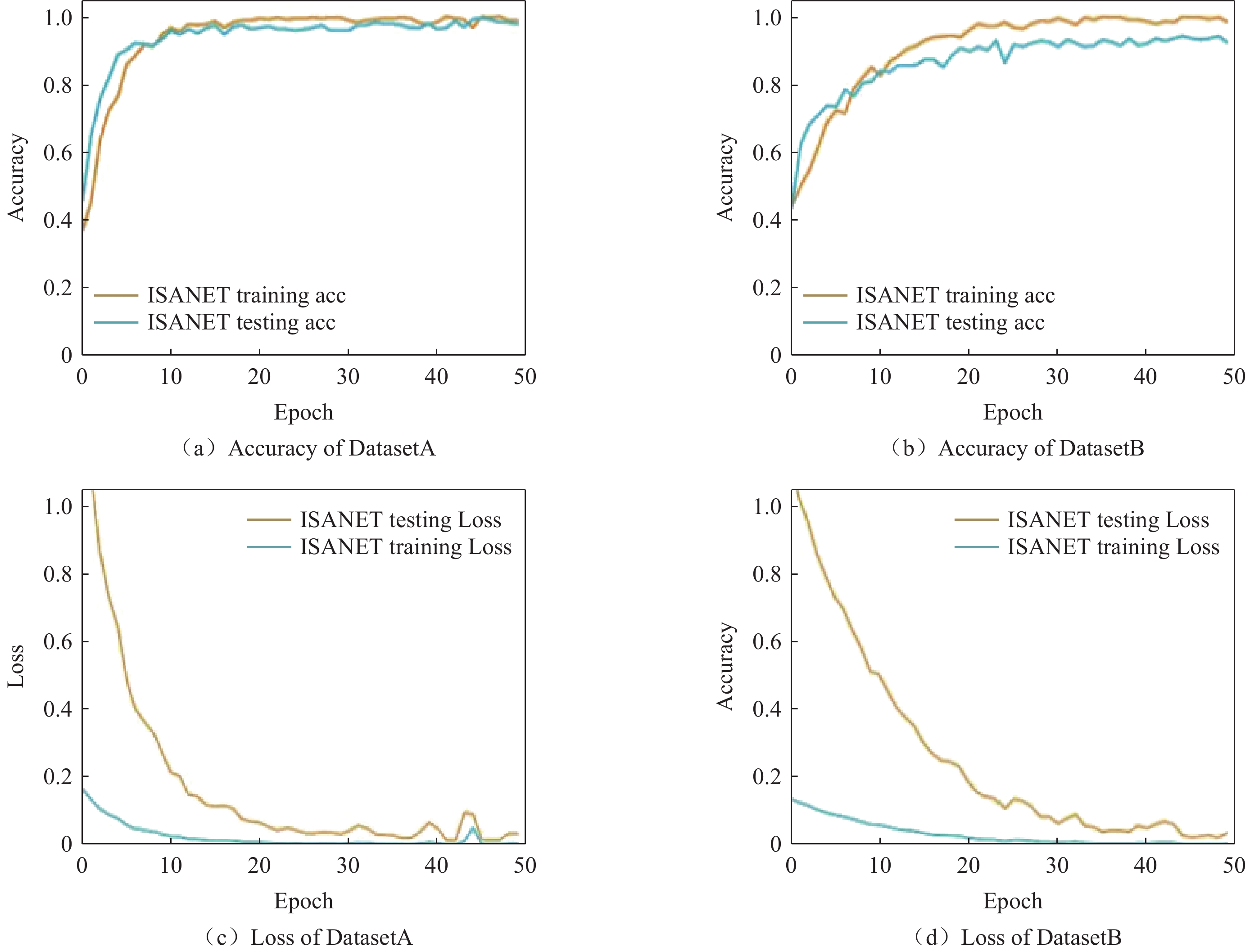
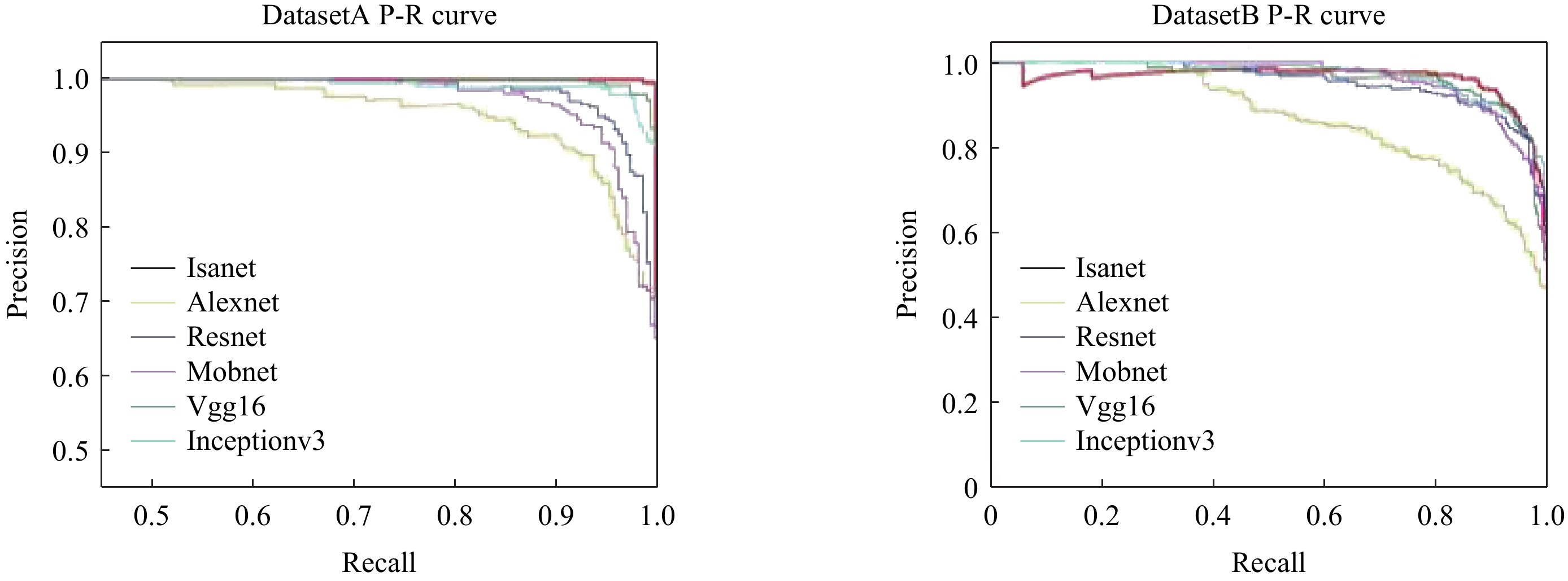
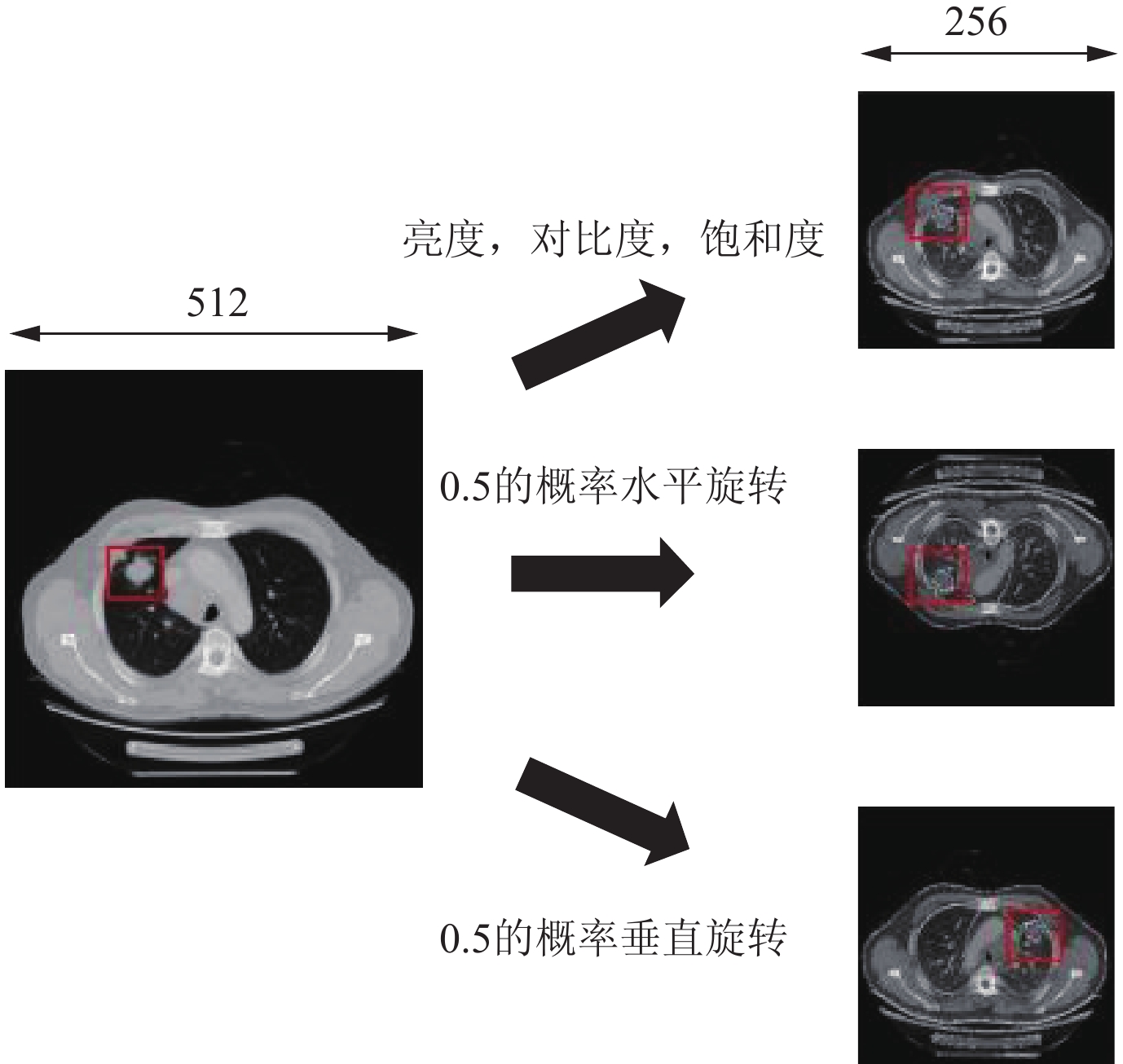
目的:构建基于卷积神经网络(CNN)和注意力机制的改进CNN模型(ISANET),评估模型的性能,并与传统CNN模型进行比较。方法:收集经手术病理证实的肺鳞癌(LUSC)或肺腺癌(LUAD)患者60例以及肺部正常患者30例的肺部CT平扫或增强图像共619张,组成DatasetA;收集公共数据集图像共737张,组成DatasetB。两数据集均按6∶4将随机分为训练集和测试集。构建ISANET模型并进行训练和验证,然后记录查准率、召回率,并计算出F1分数,用以评价ISANAT模型的效能。最后,将ISANET模型与传统CNN模型AlexNet,VGG 16,Inception V3,Mobilenet V2,ResNet 18进行对比,绘制P-R(P-R)曲线,计算出P-R曲线下面积,并评估不同模型对肺鳞癌和肺腺癌的鉴别效能。结果:相较于传统CNN模型,ISANET模型对非小细胞肺癌分类的准确度明显提高,在DatasetA和DatasetB中分别为99.6%和95.2%。结论:ISANET模型较好地实现了对肺鳞癌和肺腺癌的无创预测,提高了肺鳞癌和肺腺癌CT影像鉴别的准确度,能够帮助诊断医师对非小细胞肺癌进行快速准确的分类。
Abstract:Objective: To construct an improved deep learning computer-aided diagnosis model based on convolutional neural network (CNN) and Attention Mechanism proposed as Inception Spatial and Channel Attention Network (ISANET) and evaluate the model's performance, comparing it with the traditional CNN model. Methods: A total of 619 lung CT images of 60 patients with lung squamous cell carcinoma or lung adenocarcinoma confirmed by surgical pathology and 30 patients with normal lungs were collected retrospectively to form Dataset A; a total of 737 public dataset images were collected to form Dataset B. The two datasets were randomly divided into training and test sets at a 6:4 ratio. Construct the ISANET model and conduct training and verification, then record the precision ratio and recall ratio to calculate the F1 score and evaluate the performance of the ISANAT model. Finally, the ISANET model was compared with the traditional CNN models such as AlexNet, VGG16, InceptionV3, MobilenetV2, and ResNet18 by drawing the Precision-Recall (P-R) curve and calculating the area under the P-R curve to evaluate the classification performance of different models for LUSC and LUAD. Results: Compared with the traditional CNN model, the accuracy of the ISANET model for non-small cell lung cancer classification improved significantly, reaching 99.6% and 95.2% in Dataset A and Dataset B, respectively. Conclusions: The ISANET model provides better non-invasive prediction of LUSC and LUAD, improves the accuracy of CT imaging identification of lung squamous cell carcinoma and lung adenocarcinoma, and can help diagnosticians quickly and accurately classify non-small cell lung cancer.
-
Keywords:
- non-small cell lung cancer /
- CNN /
- attention mechanisms /
- computer-aided diagnosis
-
-
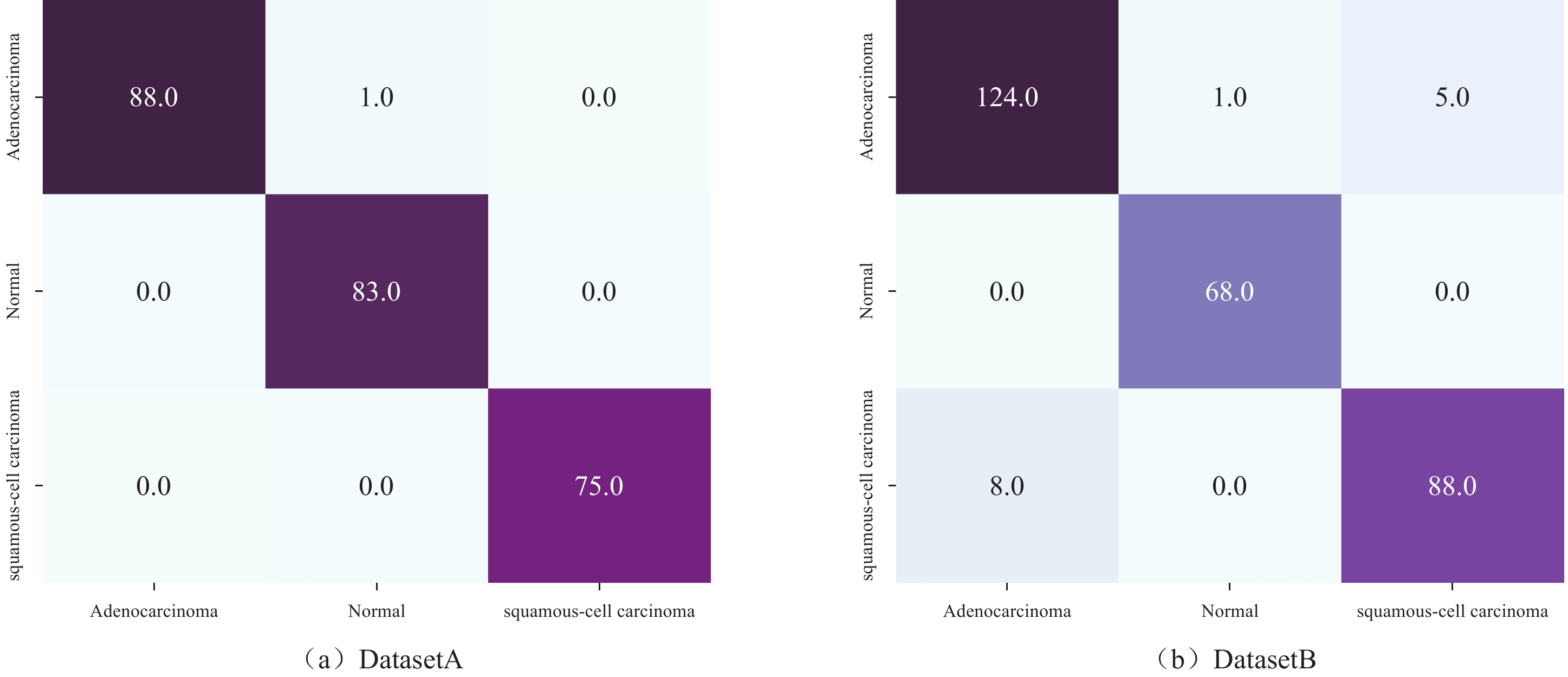
表 1 不同模型在DatasetA中的查准率、召回率和F1值比较情况
Table 1 Comparison of precision, sensitivity, and F1 score of different models in dataset A
算法名称 腺癌 鳞癌 未见明显异常 查准率 召回率 F1值 查准率 召回率 F1值 查准率 召回率 F1值 ISANET 0.898 0.925 0.906 0.946 0.914 0.928 0.959 0.985 0.968 AlexNet 0.870 0.852 0.854 0.863 0.895 0.872 0.994 0.999 0.996 VGG16 0.798 0.789 0.761 0.764 0.857 0.771 0.995 0.992 0.994 InceptionV3 0.974 0.807 0.873 0.754 0.941 0.824 0.961 0.954 0.951 MobileNetV2 0.640 0.902 0.733 0.933 0.711 0.799 0.959 0.963 0.953 ResNet18 0.919 0.929 0.919 0.927 0.905 0.911 0.960 0.992 0.969 表 2 不同模型在DatasetB中的查准率、召回率和F1值比较情况
Table 2 Comparison of precision, sensitivity, and F1 score of different models in dataset B
算法名称 腺癌 鳞癌 未见明显异常 查准率 召回率 F1值 查准率 召回率 F1值 查准率 召回率 F1值 ISANET 0.911 0.824 0.862 0.754 0.882 0.808 0.937 0.942 0.936 AlexNet 0.846 0.740 0.779 0.575 0.816 0.646 0.992 0.895 0.941 VGG16 0.814 0.720 0.717 0.507 0.668 0.531 0.964 0.949 0.952 InceptionV3 0.883 0.835 0.849 0.758 0.824 0.777 0.876 0.938 0.882 MobileNetV2 0.896 0.748 0.805 0.601 0.858 0.670 0.939 0.904 0.910 ResNet18 0.831 0.866 0.842 0.792 0.785 0.788 0.989 0.874 0.926 表 3 各模型在不同数据集中的准确度
Table 3 Accuracy of each model in different datasets
算法名称 Dataset A Dataset B ISANET 0.996 0.952 AlexNet 0.951 0.854 VGG16 0.939 0.901 InceptionV3 0.980 0.946 MobileNetV2 0.964 0.932 ResNet18 0.992 0.939 表 4 消融实验结果
Table 4 The results of ablation experiments
组别 Dataset A Dataset B A组 0.806 0.745 B组 0.810 0.735 C组 0.854 0.796 D组 0.802 0.721 -
[1] HERBST R S, MORGENSZTERN D, BOSHOFF C. The biology and management of non-small cell lung cancer[J]. Nature, 2018, 553(7689): 446-454. DOI: 10.1038/nature25183.
[2] 中华医学会肿瘤学分会, 中华医学会杂志社. 中华医学会肺癌临床诊疗指南(2022版)[J]. 中华肿瘤杂志, 2022, 44(6): 457-490. DOI: 10.3760/cma.j.cn112152-20220413-00255. ONCOLOGY SOCIETY OF CHINESE MEDICAL ASSOCIATION, CHINESE MEDICAL ASSOCIATION PUBLISHING HOUSE. Chinese Medical Association guideline for clinical diagnosis and treatment of lung cancer (2022 edition)[J]. Chinese Journal of Oncology, 2022, 44(6): 457-490. DOI: 10.3760/cma.j.cn112152-20220413-00255.
[3] CARRILLO-PEREZ F, MORALES J C, CASTILLO-SECILLA D, et al. Non-small-cell lung cancer classification via RNA-Seq and histology imaging probability fusion[J]. BMC Bioinformatics, 2021, 22(1): 454. DOI: 10.1186/s12859-021-04376-1.
[4] WOODARD G A, JONES K D, JABLONS D M. Lung cancer staging and prognosis[J]. Cancer Treatment and Research, 2016, 170: 47-75. DOI: 10.1007/978-3-319-40389-2_3.
[5] CAO B, WANG P, GU L, et al. Use of four genes in exosomes as biomarkers for the identification of lung adenocarcinoma and lung squamous cell carcinoma[J]. Oncology Letters, 2021, 21(4): 249. DOI: 10.3892/ol.2021.12510.
[6] KIM T-H, WOO S, YOON S H, et al. CT Characteristics of non–small cell lung cancer with anaplastic lymphoma kinase rearrangement: A systematic review and meta-analysis[J]. American Journal of Roentgenology, 2019, 213(5): 1059-1072. DOI: 10.2214/AJR.19.21485.
[7] 宋若晨, 褚相乐, 黄勇华, 等. 基于颅脑T1WI对比增强图像构建卷积神经网络模型鉴别肺癌与乳腺癌脑转移[J]. 中国医学影像技术, 2023, 39(7): 982-986. SONG R C, CHU X L, HUANG Y H, et al. Convolutional neural network model based on contrast-enhanced cranial T1WI for differentiating brain metastases from lung cancer or breast cancer[J]. Chinese Journal of Medical Imaging Technology, 2023, 39(7): 982-986.
[8] LITJENS G, KOOI T, BEJNORDI B E, et al. A survey on deep learning in medical image analysis[J]. Medical Image Analysis, 2017, 42: 60-88. DOI: 10.1016/j.media.2017.07.005.
[9] YAMASHITA R, NISHIO M, DO R K G, et al. Convolutional neural networks: An overview and application in radiology[J]. Insights into Imaging, 2018, 9(4): 611-629. DOI: 10.1007/s13244-018-0639-9.
[10] RUSSAKOVSKY O, DENG J, SU H, et al. ImageNet large scale visual recognition challenge[J]. International Journal of Computer Vision, 2015, 115(3): 211-252. DOI: 10.1007/s11263-015-0816-y.
[11] SZEGEDY C, VANHOUCKE V, IOFFE S, et al. Rethinking the inception architecture for computer vision[C]//2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: IEEE, 2016: 2818-2826.
[12] COUDRAY N, OCAMPO P S, SAKELLAROPOULOS T, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning[J]. Nature Medicine, 2018, 24(10): 1559-1567. DOI: 10.1038/s41591-018-0177-5.
[13] JIANG X, SHEN H. Classification of lung tissue with cystic fibrosis lung disease via deep convolutional neural networks[C]//Proceedings of the 2nd International Symposium on Image Computing and Digital Medicine. Chengdu China: ACM, 2018: 113-116.
[14] MOITRA D, KR. MANDAL R. Classification of non-small cell lung cancer using one-dimensional convolutional neural network[J]. Expert Systems with Applications, 2020, 159: 113564. DOI: 10.1016/j.eswa.2020.113564.
[15] 唐秉航, 王艳芳, 马力, 等. 基于混合注意力机制的肺结节假阳性降低[J]. CT理论与应用研究, 2022, 31(1): 63-72. DOI: 10.15953/j.ctta.2021.002. TANG B H, WANG Y F, MA L, et al. False positive reduction of pulmonary nodules based on mixed attentional raechanism[J]. CT Theory and Applications, 2022, 31(1): 63-72. DOI: 10.15953/j.ctta.2021.002.
[16] TERAMOTO A, TSUKAMOTO T, KIRIYAMA Y, et al. Automated classification of lung cancer types from cytological images using deep convolutional neural networks[J]. BioMed Research International, 2017, 2017: 1-6.
[17] TERAMOTO A, YAMADA A, KIRIYAMA Y, et al. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network[J]. Informatics in Medicine Unlocked, 2019, 16: 100205. DOI: 10.1016/j.imu.2019.100205.
[18] 樊雪林, 文昱齐, 乔志伟. 基于Transformer增强型U-net的CT图像稀疏重建与伪影抑制[J]. CT理论与应用研究(中英文), 2024, 33(1): 1-12. DOI: 10.15953/j.ctta.2023.183. FAN X L, WEN Y Q, QIAO Z W. Sparse Reconstruction of Computed Tomography Images with Transformer Enhanced U-net[J]. CT Theory and Applications, 2024, 33(1): 1-12. DOI: 10.15953/j.ctta.2023.183.
[19] KHAN M A, RAJINIKANTH V, SATAPATHY S C, et al. VGG19 network assisted joint segmentation and classification of lung nodules in CT images[J]. Diagnostics, 2021, 11(12): 2208. DOI: 10.3390/diagnostics11122208.
[20] 王媛媛, 周涛, 陆惠玲, 等. 基于集成卷积神经网络的肺部肿瘤计算机辅助诊断模型[J]. 生物医学工程学杂志, 2017, 34(4): 543-551. DOI: 10.7507/1001-5515.201607003. WANG Y Y, ZHOU T, LU H L, et al. Computer aided diagnosis model for lung tumor based on ensemble convolutional neural network[J]. Journal of Biomedical Engineering, 2017, 34(4): 543-551. DOI: 10.7507/1001-5515.201607003.
[21] AKSELROD-BALLIN A, KARLINSKY L, ALPERT S, et al. A Region Based Convolutional Network for Tumor Detection and Classification in Breast Mammography[M]//Carneiro G, Mateus D, Peter L, et al. Deep Learning and Data Labeling for Medical Applications: Vol. 10008. Cham: Springer International Publishing, 2016: 197-205.
[22] HELWAN A, EL-FAKHRI G, SASANI H, et al. Deep networks in identifying CT brain hemorrhage[J]. Journal of Intelligent & Fuzzy Systems, 2018, 35(2): 2215-2228.
[23] ESTEVA A, KUPREL B, NOVOA R A, et al. Dermatologist-level classification of skin cancer with deep neural networks[J]. Nature, 2017, 542(7639): 115-118. DOI: 10.1038/nature21056.
[24] HARANGI B. Skin lesion classification with ensembles of deep convolutional neural networks[J]. Journal of Biomedical Informatics, 2018, 86: 25-32. DOI: 10.1016/j.jbi.2018.08.006.
[25] ALOM M Z, YAKOPCIC C, NASRIN MST S, et al. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network[J]. Journal of Digital Imaging, 2019, 32(4): 605-617. DOI: 10.1007/s10278-019-00182-7.
[26] SANDLER M, HOWARD A, ZHU M, et al. MobileNetV2: Inverted residuals and linear bottlenecks[C]//2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, UT: IEEE, 2018: 4510-4520.
[27] SZEGEDY C, LIU W, JIA Y Q, et al. Going deeper with convolutions[C]//2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Boston, MA, USA: IEEE, 2015: 1-9.
[28] IOFFE S, SZEGEDY C. Batch normalization: accelerating deep network training by reducing internal covariate shift[M]. arXiv, 2015. http://arxiv.org/abs/1502.03167, 2015-03-02.
[29] 王坤, 王冠民, 路志凯. 动态增强CT对周围型肺癌的诊断价值[J]. 肿瘤研究与临床, 2021, 33(4): 304-306. DOI: 10.3760/cma.j.cn115355-20201127-00667. [30] 薛彩强, 杜晓灏, 金龙, 等. 基于MRI深度学习模型预测WHOⅡ、Ⅲ级胶质瘤MGMT启动子甲基化状态[J]. 中华放射学杂志, 2021, 55(7): 734-738. XUE C K, DU X H, JIN L, et al. Prediction of methylation status of MGMT promoter in WHO grade Ⅱ, Ⅲ glioma based on MRI deep learning model[J]. Chinese Journal of Radiology, 2021, 55(7): 734-738.
[31] 孟宇, 马之骋, 阮敬儒, 等. 基于Faster R卷积神经网络构建胸部X线片异物智能检测模型的可行性研究[J]. 中华放射学杂志, 2022, 56(12): 1359-1364. MENG Y, MA Z C, RUAN J R, et al. Feasibility of constructing the intelligent detection model for foreign bodies on chest X-ray based on Faster R-convolutionalneural network[J]. Chinese Journal of Radiology, 2022, 56(12): 1359-1364.
-
期刊类型引用(2)
1. 刘蓉辉,何磊,叶玉芳,张淑倩,陈英敏. 新型冠状病毒感染的CT影像特点及演变规律. CT理论与应用研究. 2023(05): 695-701 .  本站查看
本站查看
2. 王刚,谢浩锋,郑晓林,方学文,余芬芬,袁焕初,杜贺钦,邹玉坚. 新型冠状病毒肺炎临床与CT诊断特征的初步研究——东莞地区病例分析. CT理论与应用研究. 2020(04): 407-415 .  本站查看
本站查看
其他类型引用(0)



 下载:
下载: