Differential Diagnosis of COVID-19 and Community-acquired Pneumonia Using Different Machine Learning Methods
-
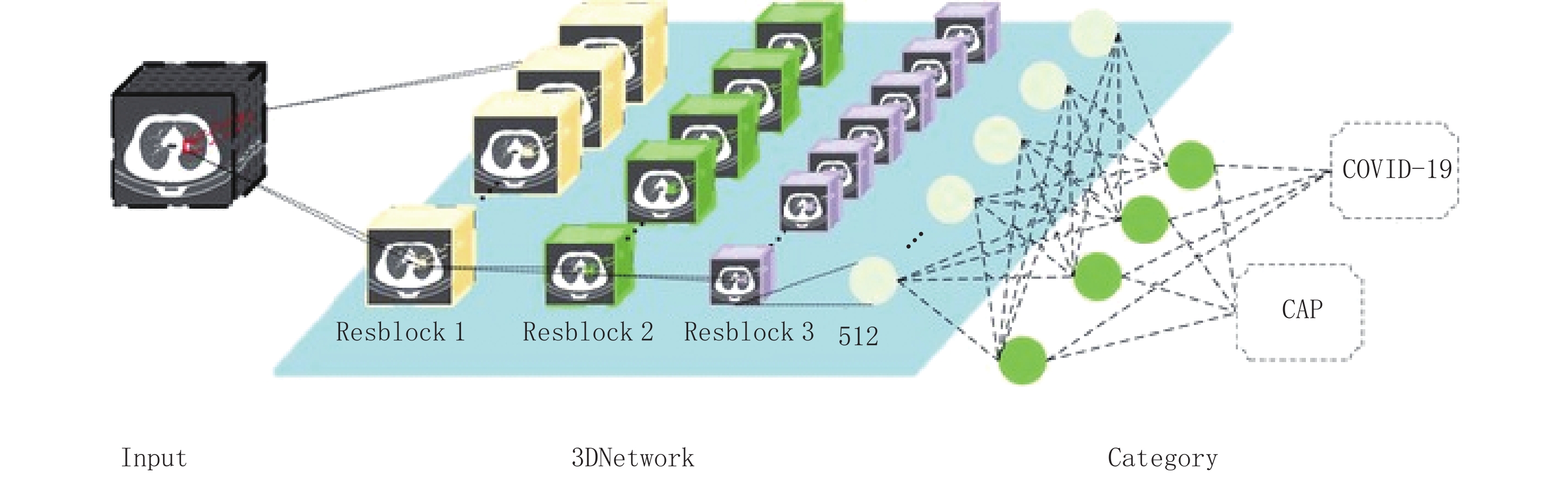
摘要: 目的:利用深度学习技术,全自动标注病变的计算机断层扫描(CT)数据,开发准确快速区分新型冠状病毒感染(COVID-19)和其他社区获得性肺炎的人工智能模型。方法:回顾性分析248例COVID-19患者及347例其他肺炎患者的资料,进行COVID-19与其他肺炎分类;在人工智能肺分割提取后将异常的CT图像特征降维,输入几种经典强化机器学习模型、三维卷积神经网络(3D CNN)和注意力多示例学习(Attention-MIL)深层神经网络架构中,模型诊断性能利用受试者工作特性(ROC)曲线、精确召回率(PR)曲线、曲线下面积(AUC)、敏感性、特异性、准确性指标进行评价。结果:在经典机器学习模型中K邻近算法(KNN)具有较好的效果,在外部测试集上的AUC值和平均精度(AP)值分别为0.79和0.89,平衡F分数(F1)值为0.76,准确率为0.75,敏感性为0.76,精确率为0.77;经典的3D CNN在外部测试集上效果良好,AUC值和AP值分别为0.64和0.82,F1值为0.71,准确率为0.78,敏感性为0.66,精确率为0.62;Attention-MIL模型在外部测试集上表现出更好的鲁棒性,AUC值和AP值分别为0.85和0.94,F1值达到0.82,准确率为0.92,敏感性为0.74,精确率为0.76。结论:与强化影像组学和3D CNN模型相比,深度学习Attention-MIL模型在鉴别诊断COVID-19和其他社区获得性肺炎上表现出更高的效能。
-
关键词:
- CT /
- Attention-MIL模型 /
- COVID-19 /
- 社区获得性肺炎
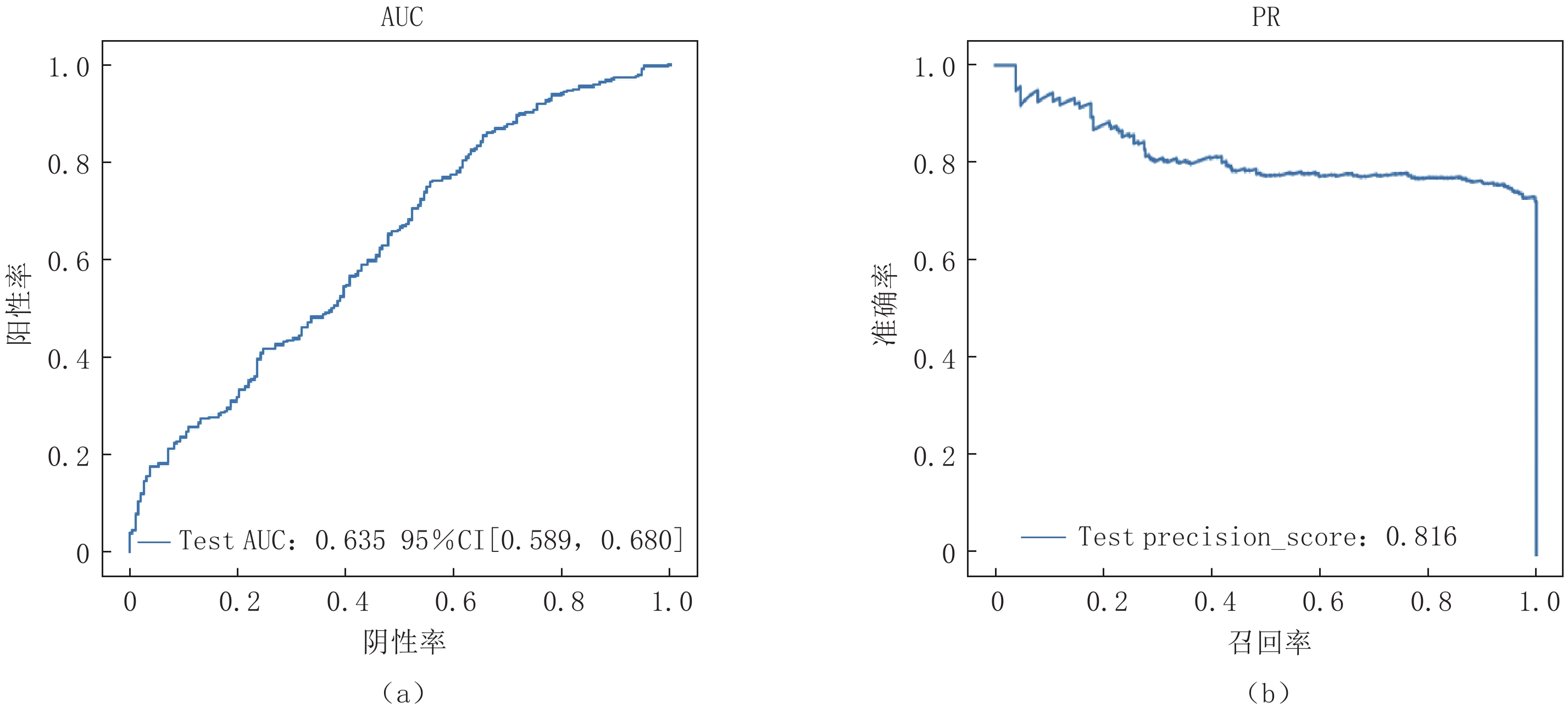
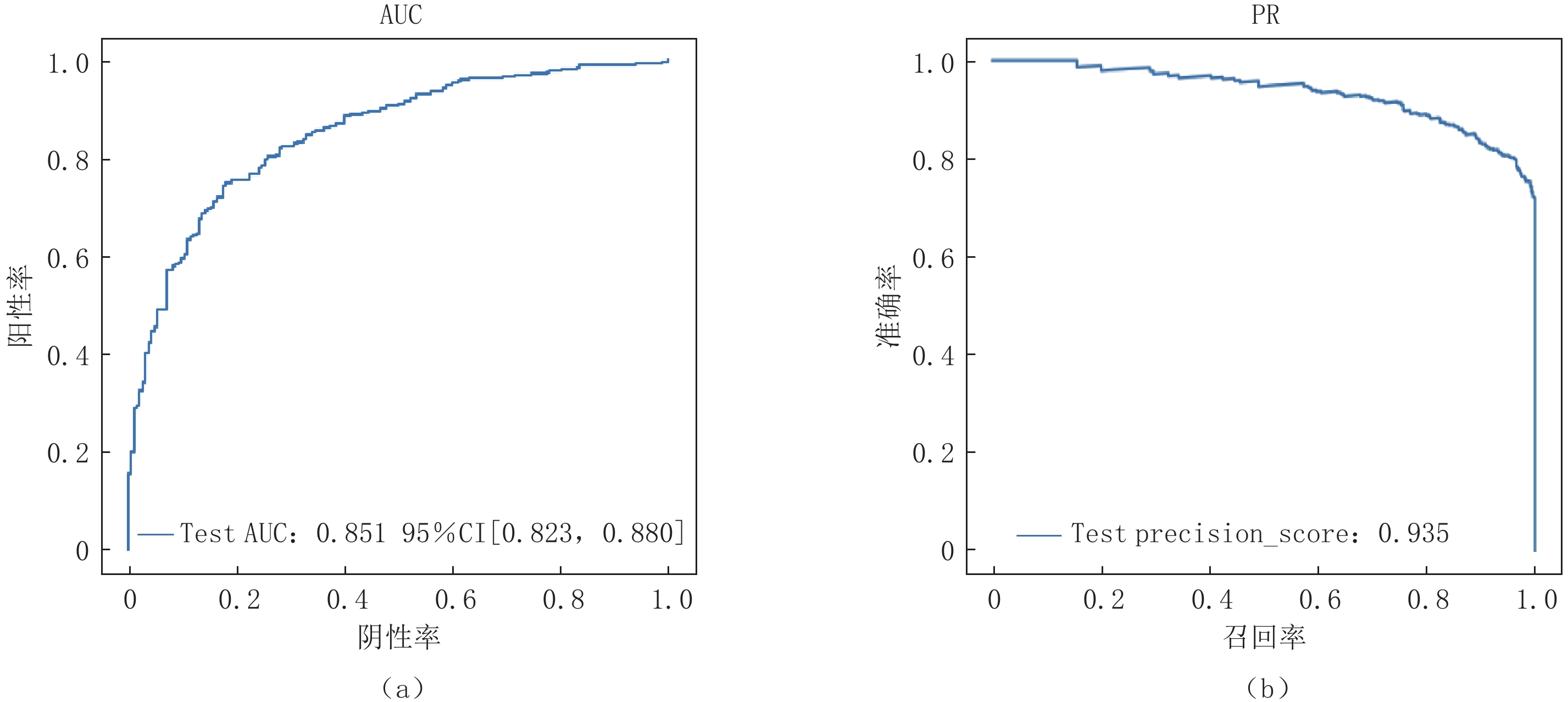
Abstract: Purpose: Utilizing deep learning techniques, this study aimed to develop an artificial intelligence model that automatically annotates lesion computed tomography (CT) data, accurately and rapidly distinguishing novel coronavirus pneumonia (COVID-19) from other community-acquired pneumonia cases. Methods: A retrospective analysis was conducted on data from 248 patients with COVID-19 and 347 patients with other types of pneumonia. The COVID-19 cases were differentiated from other pneumonia cases during classification. After performing artificial intelligence-based lung segmentation, the extracted abnormal CT image features were dimensionally reduced and inputted into various classical machine learning models, Three-dimensional convolutional neural network (3D CNN), and attention-Multiple-instance learning (MIL) deep neural network architectures. The diagnostic performance of the models was evaluated using metrics such as receiver operating characteristic (ROC) curves, Precision Recall (PR) curves, Area Under Curve (AUC), sensitivity, specificity, and accuracy. Results: Among the classical machine learning models, K-Nearest Neighbor (KNN)demonstrated good performance, with an AUC of 0.793, Average Precision (AP) of 0.886, Balanced F Score (F1-score) of 0.7608, accuracy of 0.7512, sensitivity of 0.7754, and precision of 0.7691 on the external test set. The classical 3D CNN model exhibited satisfactory performance on the external test set with an AUC of 0.635, AP of 0.816, F1-score of 0.7144, accuracy of 0.7783, sensitivity of 0.6603, and precision of 0.6200. The attention-MIL model showed better robustness on the external test set, achieving an AUC of 0.851, AP of 0.935, F1-score of 0.8193, accuracy of 0.9155, sensitivity of 0.7414, and precision of 0.7646. Conclusion: Compared to the radiomics-enhanced and 3D CNN models, the deep learning attention-MIL model exhibited better performance in the differential diagnosis of COVID-19 and other community-acquired pneumonia.-
Keywords:
- CT /
- attention-MIL model /
- COVID-19 /
- community-acquired pneumonia
-
根据2018年全球癌症统计数据,结直肠癌(colorectal cancer,CRC)是全球第2大最常见的癌症且是第4大癌症相关死亡原因[1],5年生存率仅在40%~60%。有研究表明,表皮生长因子受体(epidermal growth factor receptor,EGFR)介导的信号传导通路的异常激活与CRC患者的预后密切相关,基因突变的患者预后更差[2]。因此,肿瘤的遗传信息可作为有效的生物学指标,为患者的远期生存提供参考信息。同时,近年来通过靶向药物对患者进行个性化治疗,有效降低了死亡率,延长生存周期[3]。
RAS/RAF/MEK/ERK通路是EGFR下游信号通路中的最经典途径之一,RAS基因(如NRAS、KRAS)和BRAF基因是CRC中常见的突变基因,其中又以KRAS基因突变最为常见,约占40%[4]。美国国家综合癌症网络(National Comprehensive Cancer Network ,NCCN)指南[5]指出,RAS、BRAF等基因突变患者对靶向药物治疗缺乏反应,建议CRC患者应在治疗前明确基因型,避免靶向药物治疗无效且承受不必要的药物毒副作用。
目前,通过对活检样本或切除的组织进行病理分析来检测基因突变状态仍是最常规的做法,但有研究人员指出[6],活检样本中存在肿瘤内异质性、克隆进化和DNA质量差等情况在日常实践中价值有限。此外,循环DNA分析法也是CRC基因型检测的非侵入性方法之一,然而该方法可能因循环中的DNA不足量使得检测结果失败[7]。
因此,迫切需要一种无创且基于全肿瘤的采样技术以确定能从靶向治疗中受益的患者。非侵入式的影像学技术能从分子层面挖掘肿瘤生物学特性,不仅可以动态、全面的对CRC患者的基因状态进行分析,同时还可在患者治疗的任意阶段重复进行基因状态预测,避免活检不全面而引起的漏诊。
1. CT影像在预测CRC基因突变状态中的应用
NCCN指南推荐计算机断层扫描(CT)作为临床实践中CRC的首选成像检查,CT图像不仅可以了解肿瘤的宏观状态,同时能发掘肿瘤更深层次的特征。在Song等[8]通过分析不同CT参数及病理特征中发现,仅直肠上静脉(superior rectal vein,SRV)直径与基因突变状态较为相关,其预测KRAS突变的灵敏度(76.20%)及特异度(48.90%)均较低,因此未经处理的CT图像对于预测分子水平的生物学信息效果不佳。
令人兴奋的是,影像组学可将医学图像转换为高维特征空间,从而提高对癌症诊断与预后的预测价值。在美国国家癌症研究所(National Cancer Institute,NCI)定量成像网络(quantitative imaging network,QIN)对影像组学的大力发展下,其在肿瘤学决策方面的潜力不断增长,基于纹理的定量图像特征分析和模型建立可提供与临床诊断、实验室结果等不同的肿瘤生物学信息,有助于满足临床对CRC患者精准医疗的需求[9]。Li等[10]通过分析CRC患者术前CT图像组学特征,预测了CRC患者的KRAS基因突变状态。值得注意的是,此研究将患者临床TNM分期与CT影像组学联合,比较CRC患者单个分期与多分期组合模型的曲线下面积(area under the curve,AUC)值,结果表明单分期与CT影像学特征的联合模型可以更好地预测肿瘤特征与基因状态之间的关系(Ⅱ AUC=0.99,Ⅲ AUC=0.99,Ⅳ AUC=1.00)。
由此可见,这种临床信息与CT影像特征相结合的联合模型方式,能为提高预测效能提供新思路。
然而,随着数据科学的高速发展,机器学习作为一类新兴的研究领域,已成为影像组学不可或缺的一部分。机器学习不仅使计算机能够像人类一样学习提取及分类信息的方式,还可以自动化分析更多数据集并提取特征,进一步提高模型效能。
Yang等[11]基于机器学习的方式提取门静脉期CT图像特征,使用支持向量机(support vector machine,SVM)建立影像组学模型,结果表明:CT影像组学特征与基因突变显著相关,该预测模型的AUC值在测试集中达0.869,表现出基于机器学习的组学模型较为优异的预测效能。
深度学习(deep learning,DL)作为机器学习的子集,隐藏层神经网络模型,通过大量的向量计算,学习到数据内在规律的高阶表达特征,并利用这些特征决策的过程,能够更深入地挖掘图像中的信息,在疾病诊断、分类和预后预测等方面显示出较高效能[12]。He等[13]提取门脉期的CT图像特征,开发了基于深度学习的残差神经网络(residual neural network,ResNet)模型,结果表明仅单独瘤内特征的模型在测试队列中获得的AUC值为0.90,然而在瘤内结合瘤周组学特征时AUC高达0.930,该研究给我们带来一定启发,提取病灶影像信息不应局限于病灶本身,其周围特征也可给预测效能带来一定的提升。与此同时,肝脏作为CRC最常见的转移部位,转移病灶也包含与基因突变状态相关的特征信息,Shi等[14]使用人工神经网络(artificial neural network,ANN)方法,将临床特征(包括病变转移、单个大病变被多个小病变包绕的“微卫星状态”等)结合CT影像组学特征建立联合模型,结果表明该模型的AUC值在测试组中达0.790。
由此可见,影像组学特征与病变转移等临床特征的结合可以一定程度改善肝转移患者基因状态的预测效能。
相较于常规CT的密度测量,双源能谱CT(dual-energy spectral CT,DECT)还可提供各种参数作为定量分析的指标,其中又以碘图技术在肠道成像中效果最佳[15]。Cao等[16]通过分析DESCT参数信息结合临床危险因素开发定量诺模图以预测CRC患者的KRAS突变状态,结果表明该模型的AUC值为0.848,其决策曲线分析(decision curve analysis,DCA)也表现出预测和观察之间的良好一致性,具有可靠的临床实用性。
DESCT的多参数指标结合影像组学分析相较于常规CT成像是预测基因突变状态的更有利方法,未来或在其他疾病的诊断、预后预测等方面有更大的研究潜能。
2. MRI影像在预测CRC基因突变状态中的应用
磁共振成像(magnetic resonance imaging,MRI)作为早期CRC首选的分期工具,在肿瘤定位、切除范围及早期转移方面均具有较高的准确性,同时基于MRI影像组学在CRC的诊断分期、预后评估等表现出巨大的潜力,最新的研究表明MRI影像组学在预测基因状态方面也取得了较大的进展[17-18]。
Xu等[19]通过提取CRC患者T2加权成像(T2 WI)图像的组学特征并结合相关的临床特征进行预测CRC患者KRAS基因突变状态,结果表明联合模型的AUC值可达0.813,灵敏度为60.30%。尽管该研究所提取的MRI组学特征具有区分CRC患者KRAS基因状态的潜力,但是在提升灵敏度方面仍需要进一步研究。Oh等[20]通过提取T2 WI图像中的影像组学特征并建立决策树模型,与先前的研究相比,该研究中所使用的决策树模型的AUC值为0.884,灵敏度84.00%,在预测基因突变状态方面展现出更佳的性能。
先前的诸多MRI研究中,均通过手动分割的肿瘤区域,勾画肿瘤感兴趣区(region of interest,ROI)并提取放射组学特征,而基于深度学习的分割法有助于解决手动分割的固有局限性[21]。在Ma等[22]研究中,通过采集T2WI图像数据集,基于深度学习方法建立了一种结合空间域和频域信息的空间频率双分支注意模型(spatial frequency dual-branch attention model,SF-DBAM)预测KRAS基因的突变状态,结果表明该模型的AUC值为0.942,同时准确度及特异度分别为88.03%和90.75%,预测效能较为优异。Zhang等[23]开发了基于深度学习的3D V-net分割图像模型,提取T2WI及扩散加权成像(diffusion weighted imaging,DWI)图像中的特征,分别建立了影像组学模型,研究表明T2WI、DWI及T2WI+DWI三种模型的AUC值分别为0.714、0.816和0.887。在影像组学分析中,DWI的基因状态预测性能优于T2WI,组合方式均优于任何单一成像方式。
相较于磁共振常规序列,功能磁共振成像(functional magnetic resonance imaging,fMRI)可以根据组织成分的异质性和不规则性提供更精确的信息,广泛地运用于CRC患者肿瘤的分期、病灶转移预后评估等方面研究中。
孙丹琦等[24]研究中,同时对动态增强(DCE)图像及体素内不相干运动(intravoxel incoherent motion,IVIM)图像进行后处理,分别得到DCE及IVIM图像参数,均可在一定程度上反映肿瘤的血管化情况,然而结果表明功能参数值在组间未见显著统计学差异,功能参数与基因突变状态无显著关系,这可能是由于样本量的限制所导致,未来仍需大样本数据支持加以验证。
与之相反的是,Cui等[25]使用源于全肿瘤体积的直方图来评估扩散峰度成像(diffusion kurtosis imaging,DKI)衍生参数预测直肠腺癌患者中KRAS突变状态的潜在作用,将所有DKI相关直方图指标的诊断性能进行比较,突变型组的K相关百分位数指标高于野生型组,其中K75 th表现出最高的AUC值为0.871,灵敏度与特异度分别为81.43%和78.21%,与CRC患者的基因突变显著相关,显示出较好的预测效能。
3. 18F-FDG PET/CT影像在预测CRC基因突变状态中的应用
18F-氟代脱氧葡萄糖正电子发射断层显像(18F-deoxyglucose positron emission tomography/computer tomography,18F-FDG PET/CT)是最广泛用于肿瘤诊断的的分子成像方式。有研究表明[26],在发生基因突变的细胞中,葡萄糖转运蛋白1(glucose transporter-1,GLUT1)和葡萄糖摄取值持续增加,可能有助于无创性评估CRC患者基因突变状态。
Lovinfosse等[27]通过收集标准化摄取值(standardized uptake value,SUV)、代谢肿瘤体积和病变区域糖酵解等参数,用于评估RAS基因突变状态之间的关系,结果表明SUV与RAS基因突变呈正相关。尽管RAS基因突变的CRC患者有着更高的葡萄糖代谢具有一定的提示作用,但这可能与肿瘤患者自身的局部炎症有关,因此仅从18F-FDG PET/CT检查中提取的结直肠肿瘤的定量指标参数仍无法有效满足临床诊疗需求。
在最近的研究中,Mao等[28]将已发生肝转移的CRC患者纳入研究,在18F-FDG PET/CT检查中采取双时间点提取参数法,早期和延迟扫描中测量每个肝脏转移病灶的最大准化摄取值(max standardized uptake value,SUVmax),并且计算SUVmax差值和保留指数(retention index,RI)的变化。结果表明:延迟扫描中SUVmax与RAS基因突变状态显著正相关,其灵敏度、特异度和准确度与未发生肝转移的患者相比均有明显提高,分别为83.80%、62.00% 和71.30%。
然而部分研究[29]提出了相反的观点,研究者发现18F-FDG PET/CT中核素摄取强度与KRAS基因突变状态无显着相关性,其参数指标的整体准确性较低,无法就预测CRC基因突变状态为临床提出有价值信息。同时,最新的18F-FDG PET/CT预测CRC患者KRAS基因突变的荟萃分析[30]中也指出,18F-FDG PET/CT的合并灵敏度、特异度及诊断比值比均处于较低水平(分别为0.66、0.67、4.00),表明了18F-FDG PET/CT可能无法有效预测CRC癌患者基因突变状态。这种原因可能是因为CRC患者局部炎症反应等产生的假性参数,对结果分析产生了一定影响。因此,NCCN指南(2017)建议18F-FDG PET/CT应仅用于其他影像学检查无法确诊或可通过手术治愈的CRC患者。
4. 挑战与展望
如上述讨论,随着影像学技术的不断发展,影像组学模型在CRC中的应用已被证明效果较为优越,尤其是在加入机器学习后在CRC患者基因突变状态预测等方面取得了较大的进展,在未来还有望与人工智能(artificial Intelligence,AI)相结合,以全自动的方式对基因突变状态进行无创、实时及全面的预测将更加有效满足临床精准医疗的需求。此外,新的CT成像技术以及MR新序列均有望进一步推广应用于临床实践中,能帮助CRC患者进行更加详尽的病情判断[15]。
然而,作为影像组学的普遍局限性,不同研究中的扫描参数、特征提取、软件模式的差异,可能会使结果产生一定的偏差,因此组学模型是否可重复仍然存在不少争议。其次,目前大多数影像组学研究大多采取回顾性的研究方法,同时缺乏多中心数据的独立外部验证,均对最终的研究结果的可靠性及适用性产生影响[8]。因此作为研究者,应在模型设计阶段把握研究方向,尽可能多的收集临床及影像资料,使得研究结果的解释相对合理并能够被临床医师所接受与信赖。最后,先进的影像学技术预测CRC患者的基因突变状态已经取得一定的成效,接下来如何通过影像学方法进一步预测基因突变患者的疗效及预后情况,还有待于进一步探究。
-
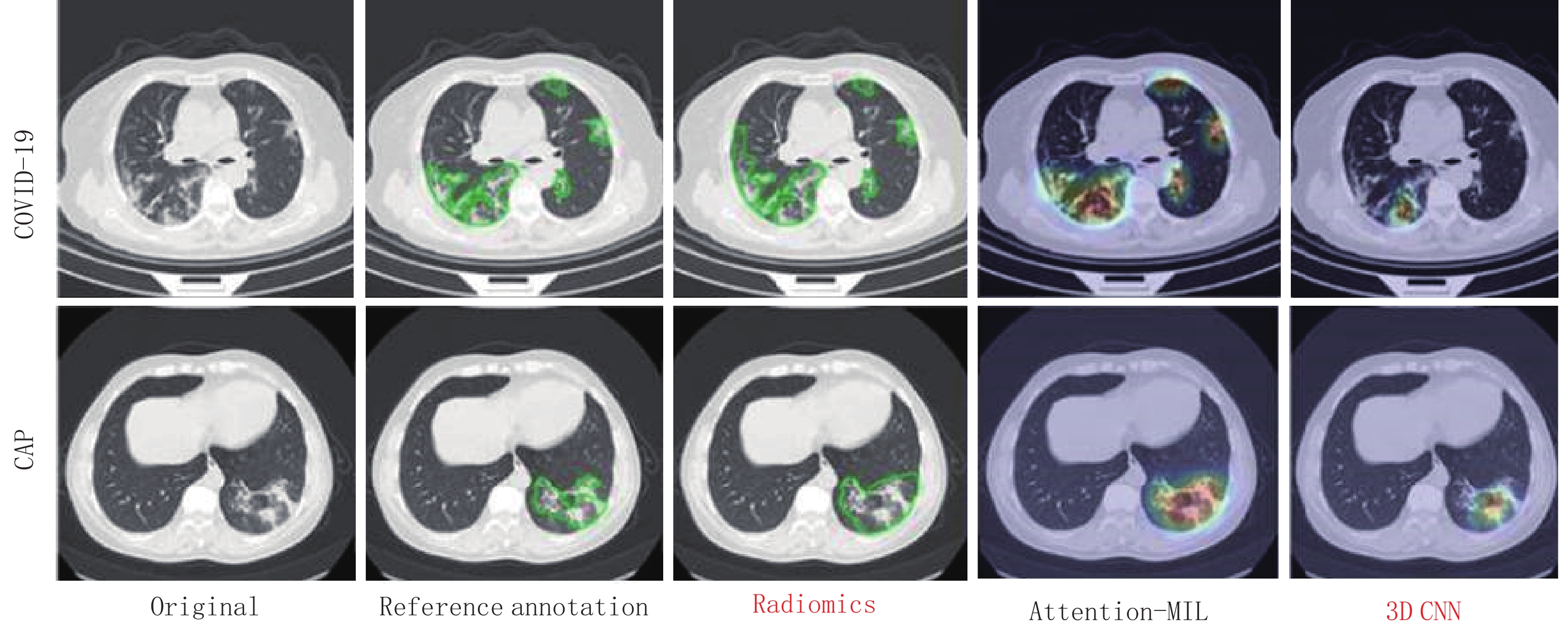
图 6 新型冠状病毒感染、社区获得性肺炎者使用梯度加权激活映射或Grad-CAM方法生成的注意力热图
热图是标准的Jet颜色图,并与原始图像重叠。红色突出显示与预测类别关联的激活区域。
Figure 6. Coronavirus disease 2019 (COVID-19), a representative example of attention heatmaps generated with data from individuals with community-acquired pneumonia using gradient-weighted category activation mapping or the Grad-CAM method-pneumonia
表 1 不同医院患者的统计数据汇总
Table 1 Summary of the statistical data of patients from different hospitals
不同医院肺炎患者 病例数(CT数)/例 年龄/岁 男/例 女/例 内蒙古人民医院 COVID-19 80 45±13.11 34~77 36 44 CAP 102 56±14.12 45~67 57 45 金门县人民医院 COVID-19 143(143) 44.95±15.12 2~86 73 70 浙江省人民医院 COVID-19 4(4) 43±13.13 26~59 1 3 CAP 35(35) 42.08±14.95 10~66 21 14 浙江大学医学院附属邵逸夫医院 COVID-19 8(8) 42.75±6.33 33~51 4 4 CAP 210(334) 44.05±16.77 15~85 103 107 台州市中心医院 COVID-19 13(13) 47.76±14.22 31~74 6 7 表 2 各种方法在外部测试集上的表现评价指标
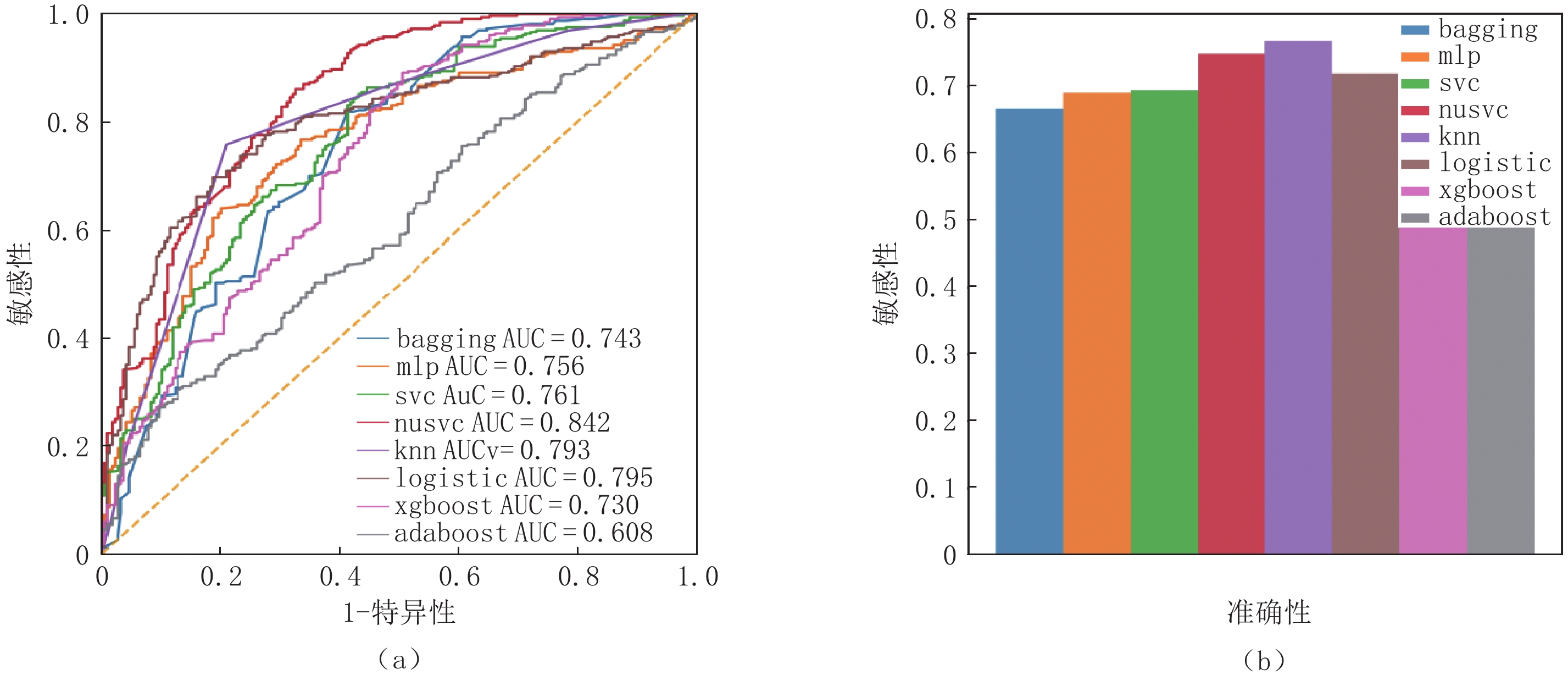
Table 2 Performance evaluation indicators for each method on independent test sets
测试集 F1值 准确率/% 召回率/% 精确率/% Adaboost 0.55 0.56 0.55 0.55 bagging 0.66 0.65 0.68 0.67 KNN 0.76 0.75 0.77 0.77 logistic 0.72 0.75 0.74 0.72 MLP 0.69 0.69 0.71 0.69 nusvc 0.74 0.75 0.76 0.75 SVC 0.68 0.69 0.68 0.69 xgboost 0.60 0.60 0.62 0.59 表 3 不同机器学习框架在COVID-19独立测试集上的性能
Table 3 Performance of different machine learning frameworks on COVID-19 independent test sets
Group/COVID-19 敏感性/% 特异性/% AUC P KNN 77 67 73 P<0.001 3D CNN 78 69 76 P<0.001 Attention-MIL 90 96 85 P<0.001 -
[1] CHEN N, ZHOU M, DONG X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study[J]. Lancet, 2020, 395(10223): 507−513. DOI: 10.1016/S0140-6736(20)30211-7.
[2] GAO Y, YAN L, HUANG Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus[J]. Science, 2020, 368(6492): 779−782. DOI: 10.1126/science.abb7498.
[3] LI Q, GUAN X, WU P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia[J]. New England Journal of Medicine, 2020, 382(13): 1199−1207. DOI: 10.1056/NEJMoa2001316.
[4] HOLSHUE M L, DEBOLT C, LINDQUIST S, et al. First case of 2019 novel coronavirus in the United States[J]. New England Journal of Medicine, 2020, 382(10): 929−936. DOI: 10.1056/NEJMoa2001191.
[5] AI T, YANG Z, HOU H, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases[J]. Radiology, 2020, 296(2): E32−E40. DOI: 10.1148/radiol.2020200642.
[6] FANG Y, ZHANG H, XIE J, et al. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR[J]. Radiology, 2020, 296(2): E115−E117. DOI: 10.1148/radiol.2020200432.
[7] 刘玉建, 仲建全, 冯浩, 等. 新型冠状病毒肺炎患者的高分辨率 CT 影像学特征[J]. 医疗装备, 2022,35(11): 1−4. LIU Y J, ZHONG J Q, FENG H, et al. Imaging characteristics of high resolution CT for patients with corona virus disease 2019[J]. Medical Equipment, 2022, 35(11): 1−4. (in Chinese).
[8] HUANG C, WANG Y, LI X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China[J]. Lancet, 2020, 395(10223): 497−506. DOI: 10.1016/S0140-6736(20)30183-5.
[9] MEI X, LEE H C, DIAO K Y, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19[J]. Nature Medicine, 2020, 26(8): 1224−1228. DOI: 10.1038/s41591-020-0931-3.
[10] CHEN Y, FAN S, CHEN Y, et al. Vessel segmentation from volumetric images: A multi-scale double-pathway network with class-balanced loss at the voxel level[J]. Medical Physics, 2021, 48(7): 3804−3814. DOI: 10.1002/mp.14934.
[11] YE H, GAO F, YIN Y, et al. Precise diagnosis of intracranial hemorrhage and subtypes using a three-dimensional joint convolutional and recurrent neural network[J]. European Radiology, 2019, 29(11): 6191−6201. DOI: 10.1007/s00330-019-06163-2.
[12] KERMANG D S, GOLDBAUM M, CAI W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning[J]. Cell, 2018, 172(5): 1122−1131.e9. DOI: 10.1016/j.cell.2018.02.010.
[13] RAJARAMAN S, CANDEMIR S, KIM I, et al. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs[J]. Applied Sciences-Basel, 2018, 8(10): 1715. DOI: 10.3390/app8101715.
[14] WYNANTS L, Van CALSTER B, COLLINS G S, et al. Prediction models for diagnosis and prognosis of COVID-19 infection: Systematic review and critical appraisal[J]. British Medical Journal, 2020, 369: m1328. DOI: 10.1136/bmj.m1328.
[15] ZHANG X, WANG D, SHAO J, et al. A deep learning integrated radiomics model for identification of coronavirus disease 2019 using computed tomography[J]. Scientific Reports, 2021, 11(1): 3938. DOI: 10.1038/s41598-021-83237-6.
[16] HUANG Y Q, LIANG C H, HE L. Preoperative prediction of lymph node metastasis in colorectal cancer[J]. Journal of Clinical Oncology, 2016, 34(18): 2157−64. DOI: 10.1200/JCO.2015.65.9128.
[17] PARMAR C, GROSSMANN P, BUSSINK J, et al. Machine learning methods for quantitative radiomic biomarkers[J]. Scientific Reports, 2015, 15: 13087. DOI: 10.1038/srep13087.
[18] NIETHAMMER M, KWITT R, VIALARD F X. Metric learning for image registration[J]. Proc EEE Computer Society Conference on Computer Vision and Pattern Recognition, 2019: 8455-8464. DOI: 10.1109/cvpr.2019.00866.
[19] ZHANG X, LU D, GAO P, et al. Survival-relevant high-risk subregion identification for glioblastoma patients: The MRI-based multiple instance learning approach[J]. European Radiology, 2020, 30(10): 5602−5610. DOI: 10.1007/s00330-020-06912-8.
[20] LIU Y, FU Q, PENG X, et al. Attention-based deep multiple-instance learning for classifying circular RNA and other long non-coding RNA[J]. Genes (Basel), 2021, 12(12): 2018. DOI: 10.3390/genes12122018.
[21] DELONG E R, DELOONG D M, CLARKE-PEARSON D L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach[J]. Biometrics, 1988, 44(3): 837−845. doi: 10.2307/2531595
[22] SELVARAJU R R, COGSWELL M, DAS A, et al. Grad-cam: Visual explanations from deep networks via gradient-based localization[J]. Proceedings of the IEEE International Conference on Computer Vision, 2017. DOI: 10.1109/ICCV.2017.74.
[23] MARTIN J, TENA N, ASUERO A G. Current state of diagnostic, screening and surveillance testing methods for COVID-19 from an analytical chemistry point of view[J]. Microchemical Journal, 2021, 167: 106305. DOI: 10.1016/j.microc.2021.106305.
[24] XU X, JIANG X, MA C, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia[J]. Engineering (Beijing), 2020, 6(10): 1122−1129. DOI: 10.1016/j.eng.2020.04.010.
[25] ABBAS A, ABDELSAMEA M, GABER M. Classification of covid-19 in chest X-ray images using DeTraC deep convolutional neural network[J]. Applied Intelligence, 2021, 51(2): 854−864. DOI: 10.1007/s10489-020-01829-7.
[26] GOZES O, FRID-ADAR M, SAGIE N, et al. Detection and analysis of COVID-19 in medical images using deep learning techniques[J]. Scientific Reports, 2021, 11(1): 19638. DOI: 10.1038/s41598-021-99015-3.
[27] CHEN J, WU L, ZHANG J, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: A prospective study[J]. Scientific Reports, 2020, 10(1): 19196. DOI: 10.1038/s41598-020-76282-0.
[28] WANG S, KANG B, MA J, et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19)[J]. European Radiology, 2021, 31(8): 6096-6104.
[29] LI Z, ZHONG Z, LI Y, et al. From community-acquired pneumonia to COVID-19: A deep learning-based method for quantitative analysis of COVID-19 on thick-section CT scans[J]. European Radiology, 2020, 30(12): 6828−6837. DOI: 10.1007/s00330-020-07042-x.
[30] CHOUAT I, ECHTIOUI A, KHEMAKHEM R, et al. COVID-19 detection in CT and CXR images using deep learning models[J]. Biogerontology, 2022, 23(1): 65−84. DOI: 10.1007/s10522-021-09946-7.
-
期刊类型引用(2)
1. 杨倩,王莉,张萍,宫艳艳,付玉叶. 基于改进Boosting集成模型的肺炎感染诊断方法. 分子影像学杂志. 2025(04): 435-440 .  百度学术
百度学术
2. 石家云,刘小峰,梁麟龙,谢齐放. 小儿支原体大叶性肺炎的诊断模型建立. 华夏医学. 2024(02): 150-155 .  百度学术
百度学术
其他类型引用(0)



 下载:
下载: