Predicting Lung Nodule Growth with Shape Transformation and Texture Learning
-
摘要:
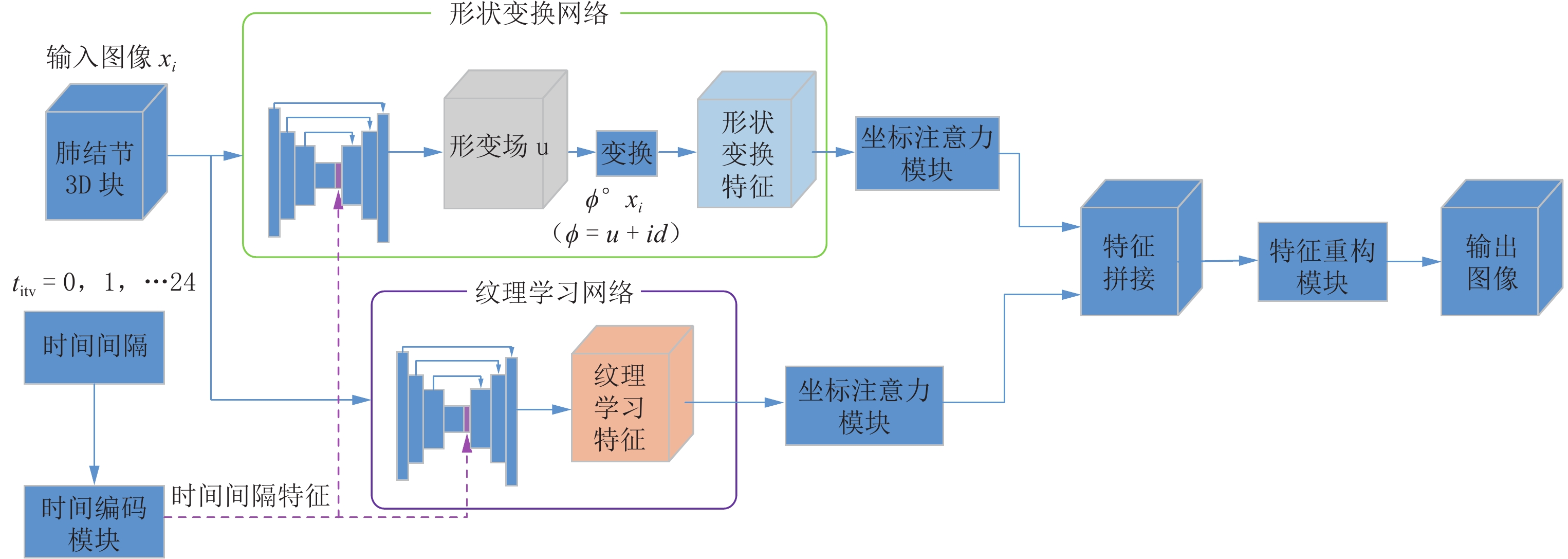
虽然人工智能在肺结节检测方面已经相当成熟,但对其生长预测的研究仍然有限。准确的生长预测有助于临床决策,为患者随访策略提供信息。本文提出一种新的结节生长预测网络模型,该模型可以在特定时间间隔生成高质量的肺结节图像。模型使用双分支结构对肺结节图像进行特征提取,其中一个分支,利用位移场预测机制,通过体素水平的未来位移估计来学习肺结节的形状转换;另一分支,采用3D U-Net,学习肺结节的纹理变化。随后,对提取的高维特征图通过坐标注意力机制,突出有利的图像特征,再拼接两个分支的结果,输入至特征重建模块得到最终的肺结节生长预测图像。同时,本文引入时间间隔编码模块,将期望的时间间隔纳入网络,从而能够生成不同未来时间点的预测图像。
Abstract:While artificial intelligence has achieved considerable maturity in lung nodule detection, research on growth prediction remains limited. Accurate growth prediction aids clinical decision-making, informing patient follow-up strategies. This paper proposes a novel nodule growth prediction network model that generates high-quality lung nodule images at specific time intervals. The model employs a two-branch structure for feature extraction. One branch, leveraging a displacement field prediction mechanism, models the shape transformation of pulmonary nodules through voxel-level future displacement estimation. The other branch, empowered by a three-dimensional U-Net, focused on learning texture changes within the nodules. A coordinate attention mechanism that emphasizes informative features within the extracted high-dimensional feature map. Subsequently, the outputs of both branches are fused and fed into the feature reconstruction module to generate the final lung nodule growth prediction image. Furthermore, a time interval coding module is introduced to incorporate the desired time interval into the network, enabling the generation of prediction images for different future time points.
-
Keywords:
- lung nodules /
- growth prediction /
- displacement field /
- time interval coding
-
-
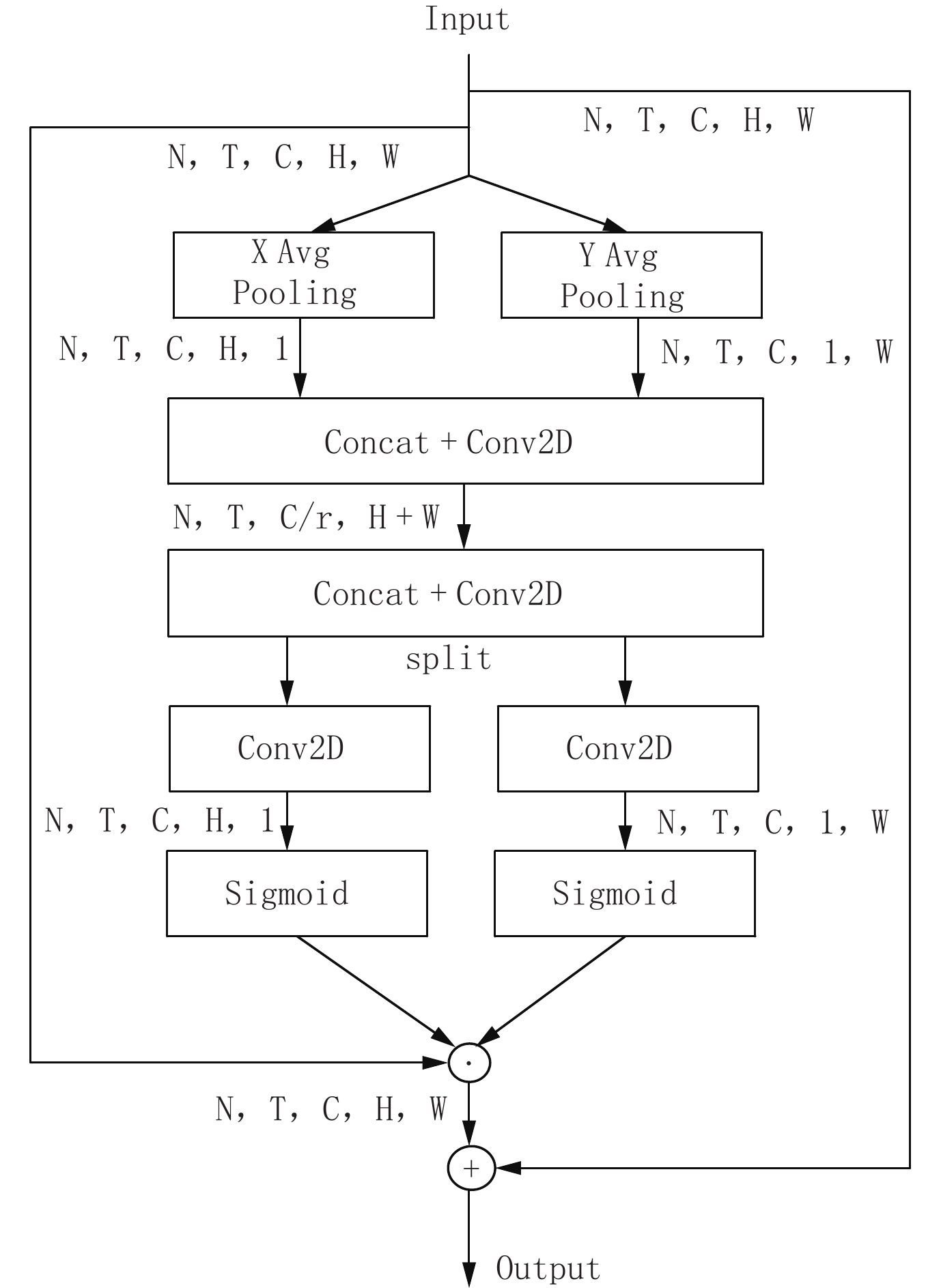
图 2 CA模块[11]
Figure 2. The coordinate attention module
表 1 实验环境
Table 1 The experimental environment
名称 配置 操作系统 Ubuntu16.04.1 LTS 编程语言 Python3.7.8 AI框架 PyTorch 1.9.0 CPU Intel(R) Core(TM) i7-7820 X CPU @ 3.60 GHz GPU NVDIA GeForce GTX 1080 Ti*2(11 GB*2) 内存 32 GB 表 2 实验结果
Table 2 The results of the experiment
方法 PSNR/dB SSIM 基线(3D U-Net) 16.12 0.8056 形状分支 18.09 0.8375 纹理分支 18.20 0.8398 形状+纹理双分支 19.31 0.8513 表 3 注意力对比实验
Table 3 The attention contrast experiment
方法 PSNR/dB SSIM SE 19.11 0.8479 CA 19.31 0.8513 表 4 同类方法对比实验
Table 4 Comparative experiment of similar methods
方法 PSNR/dB NoFoNet 18.21 本文 19.31 -
[1] OUDKERK M, LIU S, HEUVELMANS M A, et al. Lung cancer LDCT screening and mortality reduction: Evidence, pitfalls and future perspectives[J]. Nature Reviews (Clinical Oncology), 2021, 18(3): 135−151. DOI: 10.1038/s41571-020-00432-6.
[2] SHEN W, ZHOU M, YANG F, et al. Multi-scale Convolutional Neural Networks for lung nodule Classication[C]//Information Processing in Medical Imaging: 24th International Conference, UK: Springer, 2015: 588-599.
[3] SHEN W, ZHOU M, YANG F, et al. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification[J]. Pattern Recognition, 2017, 61: 663−673. DOI: 10.1016/j.patcog.2016.05.029.
[4] ZHANG L, LU L, SUMMERS R M, et al. Convolutional invasion and expansion networks for tumor growth prediction[J]. IEEE Transactions on Medical Imaging, 2018, 37(2): 638-648.
[5] RAFAEL-PALOU X, AUBANELL A, BONAVITA I, et al. Re-identification and growth detection of pulmonary nodules without image registration using 3D siamese neural networks[J]. Medical Image Analysis, 2021, 67: 101823. DOI: 10.1016/j.media.2020.101823.
[6] SHENG J, LI Y, CAO G, et al. Modeling nodule growth via spatial transformation for follow-up prediction and diagnosis[C]//2021 International Joint Conference on Neural Networks (IJCNN). Shenzhen: IEEE, 2021: 1-7.
[7] LI Y, YANG J, XU Y, et al. Learning tumor growth via follow-up volume prediction for lung nodules[C]//Proceedings of the 23th International Conference on Medical Image Computing and Computer-assisted Intervention. Peru: Springer, 2020: 508-517.
[8] RONNEBERGER O, FISCHER P, BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]//Proceedings of the 18th International Conference on Medical Image Computing and Computer-assisted Intervention. Munich: Springer, 2015: 234-241.
[9] BALAKRISHNAN G, ZHAO A, SABUNCU M R, et al. VoxelMorph: A learning framework for deformable medical image registration[J]. IEEE Transactions on Medical Imaging, 2019: 1788-1800. DOI: 10.1109/TMI.2019.2897538.
[10] HU J, SHEN L, ALBANIE S, et al. Squeeze-and-excitation networks[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2020, 42(8): 2011−2023. DOI: 10.1109/TPAMI.2019.2913372.
[11] 唐秉航, 王艳芳, 马力, 等. 基于混合注意力机制的肺结节假阳性降低[J]. CT理论与应用研究, 2022, 31(1): 63−72. DOI: 10.15953/j.ctta.2021.002. TANG B H, WANG Y F, MA L, et al. False positive reduction of pulmonary nodules based on mixed attentional mechanism[J]. CT Theory and Applications, 2022, 31(1): 63−72. DOI: 10.15953/j.ctta.2021.002. (in Chinese).
[12] HE K, ZHANG X, REN S, et al. Deep residual learning for image recognition[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016: 770-778.
[13] WANG Z, BOVIK A C, SHEIKH H R, et al. Image quality assessment: From error visibility to structural similarity[J]. IEEE Transactions on Image Processing, 2004, 13(4): 600−612. DOI: 10.1109/TIP.2003.819861.
-
期刊类型引用(2)
1. 李洋森,王伟,李炳颖,毛云新,刘晓晖. 分频AVO技术在西湖凹陷中深层薄储层评价中的应用. CT理论与应用研究(中英文). 2025(03): 409-418 .  百度学术
百度学术
2. 朱焱辉. 基于压缩感知的地震频带拓宽方法——以珠江口盆地东部惠州地区为例. 中外能源. 2023(06): 44-52 .  百度学术
百度学术
其他类型引用(0)



 下载:
下载: